
This Article From Issue
March-April 2012
Volume 100, Number 2
Page 170
DOI: 10.1511/2012.95.170
NETWORKS OF THE BRAIN. Olaf Sporns. xii + 412 pp. The MIT Press, 2011. $40.
In 1993 Francis Crick and Edward Jones published an essay in Nature titled “Backwardness of human neuroanatomy.” They lamented our poor knowledge about the anatomy and connectivity of the human brain compared to that of the macaque monkey brain, especially for the visual system. “Clearly,” they wrote, “what is needed for a modern brain anatomy is the introduction of some radically new techniques.” Networks of the Brain, by Olaf Sporns, heralds a new era in neuroanatomy based on major advances in brain imaging and brain reconstruction that have been made since Crick and Jones’s commentary nearly 20 years ago. Sporns’s goal is to connect neuroscience with network science, the study of complex networks.
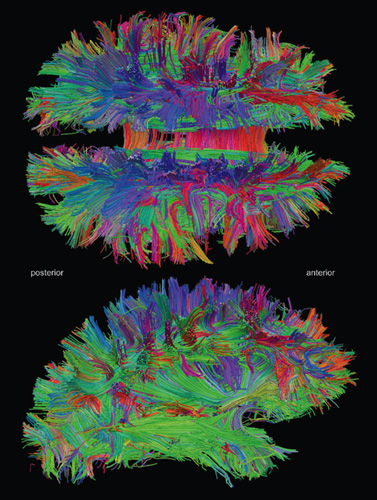
In the book’s early chapters, Sporns covers general principles of network science and offers background on the structure and dynamics of brain networks based on his research as well as that of many others, including some from my own laboratory. This prepares readers for the heart of the book, chapter 5, “Mapping Cells, Circuits, and Systems,” in which the author introduces modern imaging techniques.
Traditional techniques used to reveal brain networks are invasive and damaging; they cannot be used in humans. In neural tract tracing, for instance, a living brain is injected with chemicals to stain and reveal pathways. New imaging techniques, especially noninvasive magnetic resonance imaging (MRI), made it possible to visualize the locations of brain structures, such as the gray matter and white matter in the cerebral cortex. More recently, another breakthrough allowed researchers to trace the fibers that make up neural tracts: diffusion tensor imaging (DTI), which is based on MRI and exploits the tendency of water to diffuse more quickly along neuronal fiber bundles than it does perpendicular to them. While these techniques were being developed, computational methods for analyzing DTI data became increasingly good at producing three-dimensional maps of large-scale connectivity between brain regions, as in the image shown on the facing page. Such maps, when coupled with the detail provided by functional MRI scans, relate function to structure.
We have known for some time that there are relatively few long-range connections in the brain, and for good reason: If all 10 billion cortical neurons in the human cerebral cortex were connected with each other, the brain would be the size of a football stadium. As a consequence, the brain’s connectivity pattern is locally dense and globally sparse, following a pattern called a small-world network. The small-world effect was first documented empirically in social-network experiments by Stanley Milgram in the 1960s, and it gave rise to the idea that you are connected to anyone else in the world by no more than “six degrees of separation” through a chain of mutual acquaintances. More detailed analysis of brain structures has revealed interlocking clusters of areas that are responsible for the major brain functions, such as our sensory, motor and planning systems. In a chapter on spontaneous neural activity, Sporns details a newly discovered brain system, called the default mode, which is most active when we are being introspective rather than interacting with the world.
New techniques for reconstructing the detailed connectivity between neurons are bringing us closer to the goal of creating complete “wiring diagrams” for big brains, including those of humans. Sporns, Giulio Tononi and Rölf Kotter coined the term connectomics in 2005 to describe this new field of study. Connectomics has joined the “omic” revolution in biology, which is characterized by the collection and analysis of complete data sets for complex systems. Molecular biologist and Nobel Prize laureate Sydney Brenner was once asked which of the many omics was the most important. He replied that, in biotechnology, the only omics that really counts is economics. Indeed, economic considerations have been a driving force behind the whirlwind of data collection. The Human Genome Project first sequenced a human genome in 2001 at a cost of $3 billion. Today, the cost of sequencing a human genome is less than $10,000. The plummeting cost of computing and storing data has made it possible to collect and analyze vast databases in search of hidden patterns.
Sporns is a member of the Human Connectome Project, which was recently launched by the U.S. National Institutes of Health. The project’s goal is to produce a comprehensive structural description of all the network elements and connections in the human brain at a resolution of 1 millimeter. It is by necessity a large and expensive project: Humans exhibit a wide range of behaviors, and the sizes of brain areas vary significantly among individuals. For example, the primary visual cortex, area V1, can vary by a factor of 2 between different humans, and even between the two brain hemispheres of the same person. Similar differences have been found among individuals in the volume of white matter, which also changes in each brain during development—and in adults following learning. The Human Connectome Project will produce an invaluable quantitative treasure trove for studying the diversity of human brains.
Sporns recounts the mapping of the genome and connectome of the oft-studied roundworm Caenorhabditis elegans, whose nervous system, he notes, is “the only nervous system of any organism whose connectivity structure is completely mapped at the level of individual cells and synapses.” This was possible because the worm is only 1 millimeter long and 60 micrometers in diameter. It is extremely difficult to scale up this method to larger brains. But there is an even bigger problem: Even though neuroscientists have had the connectome of C. elegans for 25 years, the connectome in itself has not revealed how the network of neurons gives rise to behavior. To understand this, it is also essential to know the intrinsic properties of the neurons and synapses. Having a connectome alone is like having the schematic of a radio without knowing the functions of the resistors, capacitors and transistors. Creating a complete map of the human connectome would therefore be a monumental milestone but not the end of the journey to understanding how our brains work. The achievement will transform neuroscience and serve as the starting point for asking questions we could not otherwise have answered, just as having the human genome has made it possible to ask new questions about cellular and molecular systems.
Networks of the Brain explores some of these issues, such as how brain circuits develop and how individual differences in behavior relate to differences in brain circuits. But its most important contribution lies in connecting neuroscience with the science of networks. As Sporns puts it, “It’s networks all the way down.” Genes interact in gene networks, proteins in signaling networks. Information flows within and between these networks on a wide range of spatial and time scales. This is where we should be looking for solutions to the great mysteries of life and the mind.
Terrence Sejnowski is a Howard Hughes Medical Institute Investigator and the Francis Crick Professor at the Salk Institute for Biological Studies and is also a professor of biology at the University of California, San Diego. He has published over 300 scientific articles and is coauthor (with Patricia Churchland) of The Computational Brain (The MIT Press, 1992).

American Scientist Comments and Discussion
To discuss our articles or comment on them, please share them and tag American Scientist on social media platforms. Here are links to our profiles on Twitter, Facebook, and LinkedIn.
If we re-share your post, we will moderate comments/discussion following our comments policy.