The Fear of the Known
By Robert Dorit
Publishing the genetic sequence of a transmissible influenza virus might be scary, but harder decisions are yet to come.
Publishing the genetic sequence of a transmissible influenza virus might be scary, but harder decisions are yet to come.

DOI: 10.1511/2012.97.293
Spring, 1918: The war, thought to be “the war to end all wars,” had dragged on pointlessly for close to four years. The United States had only entered this conflict a year earlier, its isolationism finally overcome. By the spring of 1918, some 1.5 million U.S. servicemen had been shipped to the European theater. This number would grow to more than 4 million by war’s end. Many of these soldiers found themselves involved in trench warfare, where ground was gained, lost and regained. More than 50,000 U.S. soldiers and sailors eventually died in battle.
At about the same time, a second front, both subtle and no less deadly, was opening up far from Europe. On the morning of March 11, 1918, Albert Gitchell, a cook at Camp Funston, showed up at the camp’s infirmary complaining of a bad cold. By the end of that week, soldiers at Fort Riley, Kansas, where Camp Funston was located, were falling ill at an unusually high rate. The first deceptively simple report of what was to become one of the greatest human pandemics appeared in one sentence in the weekly journal Public Health Reports: “On March 30, 1918, the occurrence of eighteen cases of influenza of severe type, from which three deaths resulted, was reported at Haskell, Kans.” This was, for the moment, a highly contagious—but not highly lethal—strain of influenza. Most of the people afflicted by the first wave of infection would recover. But these same infected soldiers were being shipped to the European front at a rate of 10,000 a month, carrying influenza to a continent devastated by war. Placed in close quarters under unsanitary and stressful conditions, American soldiers were transporting an already potentially dangerous virus into virgin territory. Sometime between late spring and early summer of 1918, the virus evolved: It now easily infected young men and women, a cohort comparatively spared from infectious diseases. Afflicted individuals reported violent symptoms from the outset. Soon, their fevers spiked, their breathing became labored, their headaches shattering. Hours—or at most a day—later, many patients would die, drowning in their own secretions, gasping for breath.
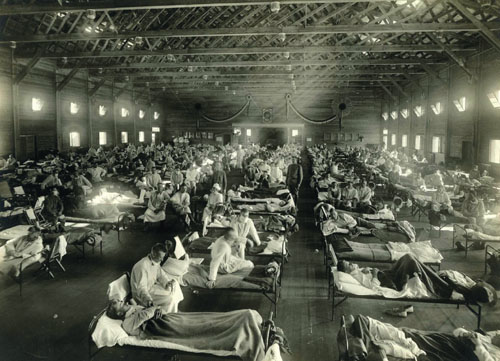
Photograph courtesy of Wikimedia Commons.
The Spanish flu pandemic of 1918 eventually claimed 57,000 American soldiers—more than died in the war itself. The movement of soldiers and civilians from infected areas disseminated this more lethal form of the influenza virus back to the United States and around the world. By the time the worldwide influenza pandemic flickered out in 1919, more than 50 million deaths could be blamed on it. Out of a world population of approximately 1.85 billion, an astonishing 25 percent may have been infected during the outbreak.
Eighty-seven years later, the first full-length portrait of the virulent 1918 influenza strain was finally completed. By piecing together fragments of the virus isolated from pathology samples preserved by the army and from the body of an Inupiat woman buried in ice for more than 75 years, scientists deciphered the full sequence of the virus.
The imminent publication of the reconstructed 1918 sequence generated considerable anticipation, but also significant anxiety. Could publication of the sequence amount to handing bioterrorists the blueprint for a lethal weapon? This issue had been carefully considered by the authors of the papers, the editors of the journals where they were ultimately published, and by a specially appointed committee of biosecurity experts (the National Science Advisory Board for Biosecurity, or NSABB). All considered the benefits of publication to far exceed the risks. Despite this process, publication was nearly thwarted by last-minute concerns from U.S. government agencies and the secretary of Health and Human Services. But on October 6, 2005, the sequence of a 1918 influenza strain was finally published in Nature, and, the following day, a companion paper in Science described the effects of the reconstructed virus on mice.
As I write, a new controversy surrounding the publication of research on the influenza virus has erupted. This time the work involves the identification of a small number of mutations in the influenza genome that affect transmissibility. Two investigators—Yoshihiro Kawaoka, from the University of Tokyo and the University of Wisconsin, and Ron Fouchier of the Erasmus Medical Center in Rotterdam—independently obtained similar results. Their papers (one of which I have not yet seen) report the creation and characterization of a strain of avian influenza (H5N1) that, in carefully controlled laboratory settings, evolved to exhibit airborne transmissibility between mammalian hosts. The experiments used ferrets as the host species, at least partially because ferrets show symptoms similar to those exhibited by humans after contracting the flu. When they get the flu, ferrets get runny noses, spike fevers, cough and, we have to assume, feel lousy. The starting point of the Imai et al. experiment was a deliberately created H5N1 strain. Influenza viruses are classified based on their two principal surface proteins: hemagglutinin (HA) and neuraminidase (NA). These two proteins define the outer appearance of the virus, and mediate the virus’s interaction with the host’s cells and its encounters with the host immune system. The H5 class HA of the starting strain meant that it was particularly effective at targeting surface receptors on the cells of birds, but comparatively inefficient at sneaking into human cells with their characteristic mammalian cell surface receptors. Taking advantage of the fragmented nature of the viral genome (more about this below), the investigators created a deliberately weakened laboratory strain of human influenza capable of expressing the H5 surface protein. Equipped only with the H5 coat protein, this strain was initially not transmissible between mammalian hosts. In order to examine whether this feeble chimera could be turned into a strain capable of ferret-to-ferret transmission, the investigators used an ingenious strategy that mimicked and accelerated the evolutionary process. To do so, they first deliberately introduced mutations in the region of the HA protein that directly interacts with host cell receptors, thus increasing the raw material on which evolution depends. They then triaged millions of viruses—each with a slightly different version of the HA protein—based on their ability to bind to engineered cells that were coated with the human form of the influenza receptor. Eight—and only eight—variants of the original strain emerged from this rigorous screen, each of them capable of binding tightly to the human form of the receptor in vitro. Eventually, only one of those variants, now placed against the genetic background of a more aggressive pandemic strain, would prove capable both of binding to the mammalian-type receptor in the lab and of successfully replicating in a deliberately infected ferret. This variant (HA N158D/N224K/Q226L/CA04) was then deliberately inoculated into the nasal cavity of ferrets, and healthy ferrets were placed in nearby cages. The naive ferrets (a description that suggests they had never seen the selected influenza strains before) could not contact or interact directly with the sickened ferrets, but the cages were designed to allow air to flow between them. Six days later, two of six naive ferrets had developed influenza: The strain had evolved the capacity to survive, reproduce and be transmitted via airborne droplets from one ferret to another. In subsequent experiments, this newly transmissible strain would acquire additional mutations that increased its infectivity and transmissibility.
These experiments showed that an H5N1 virus that was previously transmitted only between birds could, after a few generations of selection, travel between mammals in airborne droplets (such as those generated by ferret sneezes). What disconcerts is the ease with which this lock was picked: Only a handful (four to five) of recurrent mutations were necessary to account for this evolution.
Impressive as these results appear, however, context is important. Ferrets, after all, are not an exact counterpart of human beings. Like all model systems, they capture important aspects of the system we are interested in (ourselves), but they are not precise replicas. Although there can be no question about the inherent value of identifying mutations associated with increased transmissibility, we cannot assert that these ferret-adapted strains are capable of infecting humans. Also lost amid the public and regulatory anxiety surrounding these results was the observation that the lethality of these viruses was comparable to, if not lower than, that of seasonal influenza (which kills less than 0.1 percent of those it infects), a fact for which the ferrets are surely grateful. Finally, it is worth remembering that the evolved version of the HA protein resulted in successful airborne transmissibility in the context of the rest of the influenza genome in which it occurs: Other strain-specific mutations in other genes likely play a critical role in the success of this engineered virus.
In 2011, when the ferret influenza papers were submitted for publication, they triggered a review by the NSABB. This time, the review board was far from comfortable with publication. Their concern, at least initially, centered on the possibility that knowledge of the sequence changes—changes presumably tied to increased transmissibility—could be misused by enemy states or other entities to engineer a particularly threatening strain of influenza. Reconstituting a virus with the 1918 genome from scratch had always been seen as a fairly daunting task that would require considerable technical expertise. In contrast, engineering a handful of mutations into an existing avian influenza strain appeared comparatively straightforward.
In December 2011, the NSABB recommended significant changes in the manuscripts and urged that critical details of the experiments be omitted. Following a significant debate in the scientific community, the committee revisited the issue. By March of 2012, it had reversed its recommendation, arguing that the benefits of the work outweighed its potential risks. Publication has been further delayed by concerns raised (and hurdles imposed) by both the U.S. and Dutch governments. On May 2nd, the first of the papers was finally published.
To be sure, the transmissibility work, the sequencing of the 1918 influenza strain, and an increasing amount of other research in the life sciences can, in principle, be misused. Fields that deal with infectious disease, in particular, often undertake research and generate results that could be used—by agents so inclined—to design more effective biological weapons, to reduce the effectiveness of current antiviral and antibiotic therapies, or to create conditions favoring the spread of infection. But the general anxiety surrounding the publication of these latest influenza papers, much as the anxiety that surrounded the publication of the 1918 influenza genome, rests on three fundamental misunderstandings: one about the nature of the scientific enterprise, one about the character of viral evolution and one about the causes of epidemic outbreaks.
Seen from the outside, the world of professional science appears regimented and controlled. Experiments are carried out, hypotheses tested, manuscripts submitted and peer reviewed, results reproduced and confirmed. This stately flow of knowledge is certainly an important part of what we do. But coupled to it is a much more freewheeling enterprise: We converse in the elevator, send e-mails, engage in gossip around the espresso machine. While scientists can be discreet and at times secretive, we do love to talk about each other’s work. And the social structure of grant writing, conferences, posters, manuscripts, blogs and social networks means that experiments are described and discussed before, during and after they are carried out. Finally, training in science operates much like a medieval guild: Students and postdocs are mentored by one or a few individuals, and there are not many secrets in a lab.
These social trends lead directly to an inescapable conclusion: The rationale, methods and conclusions of the ferret transmissibility experiments have already been seen, heard or discussed by many individuals. While preventing publication of the manuscript would certainly limit the number of people acquainted with the results (or, more precisely, limit the rate of spread of the information), knowledge of this sort, particularly in the age of the Internet, is unlikely to be contained. Nor should it be. In the end, with very few exceptions (work that is classified from its very inception, for instance) transparency and the flow of information are central to the self-correcting nature of the scientific enterprise.
The evolutionary success of the influenza virus rests on two clever strategies: pack light and venture far. The influenza genome is a minimalist masterpiece, containing the sparest of genetic information: At 13,800 nucleotides long, its genome is roughly 100,000th the size of our own and is divided into eight RNA segments that encode 11 proteins. Like all viruses, influenza cannot replicate on its own. The information in the viral genome ensures that the virus can enter the cells of the host and, once inside, co-opt part of the machinery of the host cell for its own ends.
That light luggage is coupled with a mechanism for copying genetic information that has evolved to be remarkably … inaccurate. When influenza replicates its genetic information, it makes roughly one mistake in every 100,000 nucleotides it copies; under certain circumstances, it may make as many as one mistake for every 5,000 nucleotides. In contrast, our own replication machinery makes no more than one mistake for every 50,000,000 nucleotides copied.
The reduced fidelity of viral replication is no coincidence. Instead, it is a strategy evolved to cope with the myriad challenges facing the influenza virus, from the deadly scrutiny of the host immune system to the complex challenge of adapting to a new environment—or host. Inaccurate replication is shape-shifting at its best. As new sequences for the surface proteins of the virus result from this constant rain of mutations, these proteins take on new shapes, enabling them to evade—if only temporarily—the vigilant scrutiny of the host immune system. On occasion, two different strains meeting in the same host will take advantage of the segmented character of the influenza genome to swap entire fragments, once again giving the virus a temporary edge over the immune system.
The characteristically high mutation rate of influenza viruses has profound implications for the reports of increased transmissibility. In retrospect, the mutation rate helps explain why, after only a few rounds of selection using the ferret model system, airborne transmission evolved. The low fidelity of viral replication ensures that these mutations, and billions of other combinations of new mutations, have been—and are constantly being—generated as part of the natural history of influenza. The mutations seen in the ferret experiments are not new. They are only new to us. We had simply never seen them before in a characterized strain—possibly because in the much harsher viral world outside of the laboratory, these particular strains would not stand a chance.
Combinations of mutations that increase transmission, infectivity or virulence arise constantly. We, the human hosts, are saved by the fact that the viral lifestyle involves a set of competing claims: Mutations that increase transmissibility may inadvertently reduce the virus’s ability to enter host cells. Seen in this light, the influenza genome represents a compromise solution to a set of sometimes contradictory challenges. We are also helped by the virus’s complete dependence on its host. Influenza viruses cannot simply invade our bodies and promptly kill us off in an orgy of viral replication—that would be suicide. Instead, viruses must manage their hosts. A less transmissible strain that keeps its host alive, mobile and capable of interacting with other naive hosts may well outcompete a virus that is highly transmissible, aggressively infective and fulminating in its consequences.
Viral outbreaks are by definition the result of the interplay among virus, host and a variety of environmental, demographic and social factors. As a result, we cannot really say that the 1918 genome explains the pandemic any more than we can say that the murder of Archduke Ferdinand, by triggering the war, accounts for the pandemic. Unquestionably, certain strains of influenza are more dangerous than others: The 1918 strain made people far from the trenches in Belgium and France very ill. But the fate of even the most virulent of strains is not written solely in its genes.
In all likelihood, enough has changed over the past several decades that the 1918 strain would no longer cause a worldwide pandemic were it released today. Today’s population has been exposed to related (but less harmful) influenza strains, providing a measure of immunity. Further immunity derives from the nearly 450 million doses of flu vaccine administered worldwide annually. The advent of antibiotics would significantly reduce the development of secondary infections, one of the leading causes of influenza-related mortality in 1918. These and other changes in the overall health and nutritional status of the human population have altered the landscape of potential epidemics in radical ways. This is in no way meant to suggest that the human population in its current status is not fertile soil for pandemics. As recent experiences with severe acute respiratory syndrome (SARS) and H5N1 influenza suggest, emerging infectious diseases are constantly smoldering beneath the surface, just waiting to break out into full-fledged epidemics.
Seen in this light, the publication of the sequence changes resulting in mammal-to-mammal transmission (or of the sequence of a 1918 strain) can no longer be considered a reckless or naive act. Rather than being alarmed about the potential misuse of this research, we should be paying more attention to the detection and worldwide monitoring of new strains capable of holding their own against existing strains, and to strains exhibiting surface characteristics unfamiliar to the immune systems of much of the human population. And we should only really worry when such strains gain a foothold in parts of the world where poverty, malnutrition or unrest preclude an effective public health response.
The Kawaoka paper has now been published, and the Fouchier paper cannot be far behind. This is the proper outcome. These findings are dispatches from deep within enemy territory. The information they provide enhances our understanding of a fearsome rival and provides new targets for drug and vaccine development, as well as enhancing our monitoring capabilities. This controversy, in the end, is not really about the risk of disseminating these scientific findings. What this episode has done instead is uncover deficiencies in our ability to classify, monitor and regulate so-called dual-use research—legitimate studies whose results could also threaten public health and security. And this problem will only grow more acute. As our ability to manipulate and customize life forms using experimental evolution, brute-force screening or synthetic biology improves, we will be facing more—and more complicated—dilemmas about what research can and should be done. I cannot easily predict what form this monitoring will take, although I suspect it will be a combination of self-policing by scientists and more rigorous scrutiny of research proposals by granting panels and biosafety committees. But for now, the genie is out of the bottle, and any solution predicated on shoehorning him back in will inevitably fail.
Click "American Scientist" to access home page
American Scientist Comments and Discussion
To discuss our articles or comment on them, please share them and tag American Scientist on social media platforms. Here are links to our profiles on Twitter, Facebook, and LinkedIn.
If we re-share your post, we will moderate comments/discussion following our comments policy.