Making Life from Scratch
By Robert Dorit
Artificial intelligence is not human intelligence, nor is synthetic life the same as life with evolutionary history.
Artificial intelligence is not human intelligence, nor is synthetic life the same as life with evolutionary history.

DOI: 10.1511/2013.104.342
On May 11, 1997, Garry Kasparov pushed his chair back from the table and walked away from the chessboard. After only 19 moves, he conceded the game to Deep Blue, a massive parallel-processing computer built and programmed by IBM. For the first time ever, a machine had succeeded in defeating a chess Grand Master.

Tom Mihalek/AFP/Getty Images
Around the world, headlines declared the victory of machine over man. Some proclaimed that a computer was now as smart as a human being. Although the outcome was most certainly a landmark in the evolution of computers and in the field of artificial intelligence, many commentators missed what was perhaps the most profound lesson of that encounter. Deep Blue indeed managed to defeat Kasparov, but only by doing what computers do best: crunching numbers. Some reporters spoke anthropomorphically about Deep Blue’s “strategy,” but as strategies go, it was remarkably lacking in finesse. Deep Blue mindlessly analyzed virtually every conceivable play and its consequences several moves down the line. After that, the machine countered Kasparov according to predetermined criteria.
Deep Blue’s display of computational brute force had nothing to do with the way Kasparov (or even I, for that matter) plays chess. Far from being a defeat for human intelligence, the outcome demonstrated that Kasparov could, through training and intuition, play a beautifully thoughtful game of chess, whereas a computer could play only crudely and mechanically—through exhaustive calculation. It had taken IBM 12 years of concerted work to build a machine powerful enough (not smart enough) to defeat Kasparov. Even so, Kasparov was demonstrably rattled by the result. He should not have been.
I do not mean to downplay the significance of Deep Blue’s win. Tremendous human ingenuity went into developing its hardware and software. But the route to Deep Blue’s victory underscores the fact that artificial intelligence, although compelling in its own right, is not a silicon-based version of human intelligence. When Deep Blue won at chess, the machine did so precisely by not mimicking the operation of the human brain. Deep Blue did not triumph by being intelligent. It triumphed by being big.
Today, a new area of research that similarly aims to mimic a complex biological phenomenon—life itself—is taking off. Synthetic biology, a seductive experimental subfield in the life sciences, seems tantalizingly to promise custom-designed life created in the laboratory. But here, too, I suggest that language—word choice fueled by an ambitious agenda—misleads us. Just as Deep Blue shed some light on, but did not replicate, the workings of Kasparov’s brain, synthetic biology imitates but cannot replicate living organisms.
Today, the prospect of understanding life by making life is no longer untenable.
Fueled in part by the triumphs of reductionist biology—the deciphering of the fundamental molecular mechanisms of life, the full cataloguing of chemical reactions in the cell, and the decoding of genomes—researchers are seeking not just to understand life but also to make organisms that will do our bidding: make biofuels, synthesize pharmaceuticals, make chemicals sustainably, clean fouled water, produce food, and fight disease. Technological breakthroughs, including the automation of laboratory tasks and dramatic increases in computational storage and power (comparisons with Deep Blue beckon) have expanded the scope of synthetic biology. In 2000 the first synthetic genomes, assembled by “cutting and pasting” sequences from existing genomes and then parsing them together, were successfully inserted into Escherichia coli. In 2008 researchers at the J. Craig Venter Institute announced the successful synthesis of the smallest bacterial genome, that of Mycoplasma genitalium, over half a million bases in length. In 2010 a one-megabase synthetic Mycoplasma mycoides genome was inserted into a Mycoplasma capricolum recipient cell, effectively transforming one Mycoplasma species into another in a single stroke.
Today, the prospect of understanding life by making life is no longer untenable. The goals of synthetic biology are Promethean, and I suspect that both our failures and our probable eventual successes will indeed end up teaching us about the actual world of the living—just not in the ways we might expect.
Trading on the multiple meanings of the word synthetic, two radically different approaches to synthetic biology have emerged, each of which embodies different assumptions about the living world and about the goals of the field.
Barbara Aulicino
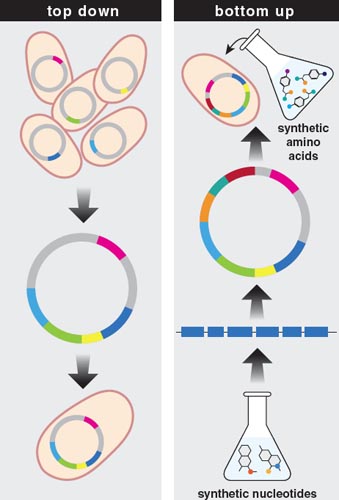
The first of these approaches, sometimes called bottom-up or de novo, seeks to synthesize new kinds of cells from scratch. This strategy addresses a profound—literally, a fundamental—question in biology: To what extent is the history of life on this or any other planet constrained by the chemistry of the building blocks of life?
For a long time, questions about alternative unfoldings of the history of life were no more than the stuff of late-night musings at scientific meetings. But my colleagues and I knew the questions to be profound and not merely abstract. Over the past two decades, scientists have begun seriously investigating alternative chemistries and organizations for living systems on other planets. This effort has gained importance as astronomers have intensified the search for life on Mars, and as the estimate of potentially habitable planets in the Solar System and beyond continues to expand. If the only life known (that on Earth) is but one of many conceivable ways of building what is called a cell—a system capable of evolving—then scientists risk overlooking life forms that do not exhibit the signatures of Earthlike life.
Barbara Aulicino
In the past, my colleagues were confined to raising such issues without being able to address them experimentally. After all, life reflects its single common origin by relying on nucleic acids (RNA and DNA) for the preservation and transmission of information and on proteins for most of its essential molecular functions. Everything, in every cell of every organism on this planet, evolved in response to these fundamentals. There were quite literally no alternatives on Earth to study.
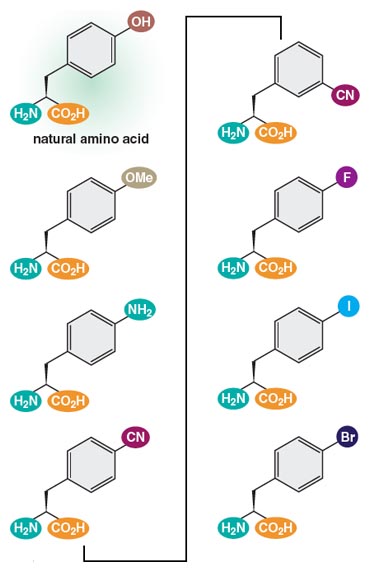
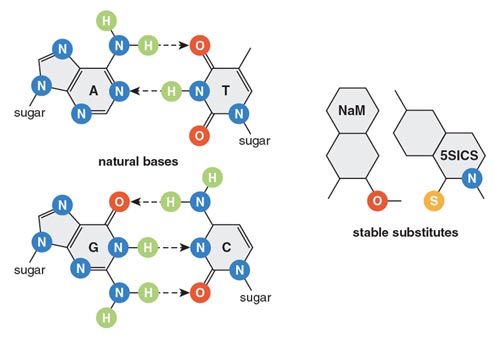
The lack of points of comparison did not daunt us, however. Instead, a number of labs decided to address the question by chemically synthesizing novel building blocks—modified amino acids and new nucleotides—and exploring how these new chemistries might throw open previously closed evolutionary pathways. This synthesis approach proved to be no simple undertaking: Aberrant amino acids and nucleotides occasionally arise spontaneously in living cells, but they are always bad news for a healthy cell because they can cause misfolding of proteins or stop replication in its tracks. As a result, multiple systems have evolved in all organisms precisely to guard against the incorporation of such novel building blocks into the cell’s nucleic acids and proteins. But over the past decade, experimental imagination and persistence have enabled investigators to select variants of existing molecular machinery that can work with these new building blocks. In so doing, my colleagues have succeeded in adding new text to that Rosetta Stone of living systems, the genetic code. New or modified nucleotides expand the canonical A–T and G–C DNA pairings, thus increasing the potential number and nuance of hereditary information. For instance, two novel nucleotides, called NaM and 5SICS, have recently been shown to pair (albeit in an unusual way) within the DNA double helix and can be replicated by the cell’s machinery. These novel nucleotides have novel geometries and hence differ slightly in how they pair with their counterparts across the DNA double helix. Similarly, the expansion of the amino acid vocabulary beyond the 20 or so amino acids normally encountered in living organisms, in turn, makes possible the synthesis of proteins with novel stabilities, architectures, and functions.
Barbara Aulicino
These are early days still, but new building blocks with unique properties continue to be synthesized. At the same time, they remain compatible with the roles played by existing nucleic acids and proteins. These results already make one thing clear: The world of naturally occurring genetics here on Earth is certainly not the only possible world.
A second approach in synthetic biology, the top-down strategy, aims to create customized organisms from a growing catalog of existing molecular parts. Proponents of this second approach argue that biologists have finally come to describe and understand living systems in sufficient detail that they can now customize those systems to create specific desired outcomes and address specific human needs. I call this the “LEGO view” of the world. Here, the genome is regarded simply as the sum of its parts. These parts are, in effect, modular, and supposedly retain their integrity and function independent of the context in which they occur. If this were so, biological engineers would be able to select desired genes, assemble them into a genome and, in so doing, give rise to an organism that might never have evolved on its own.
Modularity, particularly at the molecular level, seems at first to be quite common in the living world and underlies the entire biotechnology revolution. Modularity, after all, enabled early practitioners to transfer a human gene into the genome of a bacterium and still have it direct the the production of a desired human protein. Nature, too, often trades in modular subunits: Protein domains, individual genes, or clusters of genes that confer particular phenotypes (for example, resistance to multiple antibiotics) have moved from one location within a genome to another, or from one genome to another, all the while retaining their integrity and function. For example, the well-known Hox genes, responsible for controlling embryonic development along a central axis, have undergone repeated expansions, contractions, duplications, and changes of address within eukaryotic genomes while retaining their fundamental role: determining crucial patterns of early development in multicellular animals. And even larger genome segments spanning thousands of genes have, over evolutionary time, moved from one location to another without losing their function.
A short leap separates these observations from the conviction that all genetic information is fundamentally modular, and so can be disassembled and reassembled in virtually infinite combinations. As the catalog of known genetic elements continues to grow, the vision of creating organisms that will do exactly what researchers want them to do beguiles. Synthetic biology promises that the power of the living world, fully dissected and deconstructed in the first triumphant half-century of the Age of Biology, will finally be brought to bear in solving humanity’s most complex and recalcitrant problems, from the production of virtually inexhaustible food supplies to the waste-free transformation of sunlight into usable energy.
Despite our growing knowledge of cellular circuitry, engineering novel organisms from existing genetic elements with the use of the top-down strategy has proven surprisingly hard. Individual genes drawn from one bacterium do not always behave as expected when placed in a different host. The more heterogeneous the collection of novel elements introduced, the less likely they are to perform as expected. Regulation, synchronization, and feedback collapse as the subtle conversation underlying all the networks in living systems degenerates into babble.
A truly synthetic cell will be a cell without ancestry, and it will teach us a great deal about the underlying complexities of life.
My colleagues and I wrestle with the seeming contradiction between the apparent modularity of genetic information and the maddeningly difficult task of designing a new organism from preexisting and well-characterized genetic information. The explanation, I suspect, lies in one of the fundamental differences between biology and engineering: the importance of history.
When researchers combine genetic information from disparate sources into a single genome, they are by definition dismissing history. In so doing, they are foolishly expecting genetic elements to behave predictably independent of their surroundings. But virtually no gene is an island, and all genetic information has been shaped by evolution in the context of an entire genome, itself harbored within an organism. As a result, only a handful of genetic elements really do behave independent of their location. Antibiotic resistance genes may be an example: Actively traded among species in the microbial world, they seem strangely impervious to their surroundings—but only because they have evolved to be so.
Evolution is a blind, brute-force experiment, testing innumerable solutions, much like Deep Blue’s playing chess by methodically examining virtually every possible move. This natural experiment, ongoing for more than three billion years, is constantly placing functional genetic elements in novel contexts. Sometimes this relocation is the result of an orderly evolved mechanism (such as recombination during cell division), sometimes it is the consequence of unexpected random events (such as breaks in a chromosome or translocation of genes across species barriers).
In evolution’s natural experiment every movement of genetic information into a new neighborhood is carefully scrutinized by selection. Genetic elements placed in new contexts coevolve with their neighbors into higher-order networks that contribute to the survival and reproduction of the organism. Those that do not are quickly weeded out by selection. This history of coevolution—of genetic elements having evolved together in the same genetic neighborhood—is what synthetic biology will need to incorporate into its approach if it hopes to realize its stated ambitions.
Similarly, practitioners of the bottom-up strategy have to contend with the myriad ways organisms have evolved to operate with the amino acids and nucleotides that were exploited—in what may have been only a fortunate accident—early in the origins of life. The entire history of life—from its beginnings as organized chemistry—has given rise to enzymes and organelles that are exquisitely dependent on the chemistry and architecture of the existing nucleotide and amino acid building blocks. The complex interlocking cellular machinery of life evolved around this small subset of available (or easily produced) building blocks and simultaneously learned to discriminate against variants and alternatives. As a result, synthetic biologists seeking to expand the size and meaning of the genetic alphabet are constantly contending with history. Other chemical beginnings can be envisioned and synthesized, but the contingent history of life as it has actually unfolded cannot be so easily overcome, and it constrains what synthetic biology can hope to achieve with new fundamental blocks.
Our efforts in synthetic biology have already taught us a great deal. By confirming the fact that life as it exists today is but one realized embodiment amid many possible and by reminding us yet again that contingency matters, the research agenda of synthetic biology has already borne fruit. I suspect that we may eventually succeed in creating an artificial cell from novel starting materials. This truly new synthetic cell may be able to sustain a complex network of coupled metabolic reactions and may even be capable of reproducing and evolving. But I would wager that it will do so in ways radically different from existing living cells.
Deep Blue taught us a great deal about the power of the human mind precisely because it could not reproduce the intuitive and logical leaps of Kasparov’s mind. A truly synthetic cell, built from scratch or even from preexisting components, will be a cell without ancestry, and it, too, will teach us a great deal about the underlying complexities of life without actually reproducing them. In so doing, it will remind us yet again that biology is not just engineering, that organisms are not just machines assembled from preexisting parts, and that history matters to living systems in profound and inescapable ways.
Click "American Scientist" to access home page
American Scientist Comments and Discussion
To discuss our articles or comment on them, please share them and tag American Scientist on social media platforms. Here are links to our profiles on Twitter, Facebook, and LinkedIn.
If we re-share your post, we will moderate comments/discussion following our comments policy.