Protein World Atlas
By Christopher Brodie
Pretty pictures mark proteins' province
Pretty pictures mark proteins' province

DOI: 10.1511/2006.57.20
If DNA is the blueprint of biology, then proteins are the general contractors, workers, power tools and half of the building materials. Yet the methods for studying proteins remain cumbersome even as genomic tools become faster and more powerful.
A team of scientists from Sweden aim to fill that gap with an ambitious project: a high-throughput, antibody-based portrait of the human proteome (the set of all proteins made by human beings). The first fruit of their labor is the freely available Human Protein Atlas, initially released in August 2005, which describes the location and quantity of about 700 proteins in 48 tissues and 20 types of cancer. The project, led by Mathias Uhlén and based at the Royal Institute of Technology in Stockholm and Uppsala University, is planned eventually to include nearly every human protein.
The atlas (www.proteinatlas.org) consists of photomicrographs made with a technique called immunohistochemistry, in which an antibody labels its target, or epitope, on a thin slice of tissue (or in this case, a "tissue microarray" of up to 1,000 samples at a time). This method pinpoints the protein to a specific compartment of a specific cell in a specific tissue. More than 500 high-resolution, high-magnification images are available for each antibody.
Most of the antibodies are generated from PrESTs, or protein epitope signature tags, that are made in vitro from predicted coding regions in the human genome. The purified "monospecific" antibodies are rigorously screened for sensitivity and selectivity. Some PrESTs correspond to well-studied genes, including ones implicated in cancer and other diseases. But others are more mysterious, representing genes that have never been investigated.
Human Protein Atlas
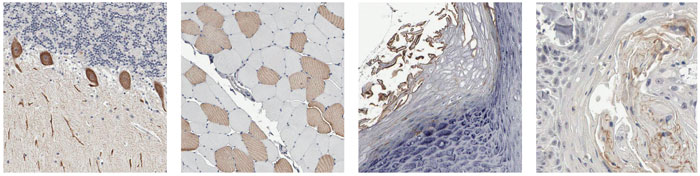
For example, the unnamed gene NP_443138.1 on chromosome 22 encodes a protein with unknown function and no clear relatives in other species, but its pattern of distribution is fascinating. Above right is the standard summary of staining intensity for this protein in various tissues and tumors. Red indicates the highest concentration of the protein, and orange and yellow signify lesser amounts. White shows a negative result; black denotes missing images. A small sample of the stained tissue sections is shown at left, in which brown marks the presence of the NP_443138.1 protein and blue is a general stain used to see the surrounding tissue. The protein is found in (from left to right) Purkinje cells in the cerebellum, skeletal muscle fibers, the outer layer of skin and subpopulations of tumor cells in cancers of the head and neck.
Describing protein location is only the first step. The project's eventual goal is to use the antibodies as tools to explore the structures, functions and interactions of proteins. Skål!
Click "American Scientist" to access home page
American Scientist Comments and Discussion
To discuss our articles or comment on them, please share them and tag American Scientist on social media platforms. Here are links to our profiles on Twitter, Facebook, and LinkedIn.
If we re-share your post, we will moderate comments/discussion following our comments policy.