Blood in Motion
By Catherine Clabby
Applied mathematician George Karniadakis models how diseases alter the body’s circulation.
Applied mathematician George Karniadakis models how diseases alter the body’s circulation.

DOI: 10.1511/2013.104.386
Precisely describing how blood flows through the smallest vessels is no simple matter. Fluid dynamics comes into play, but so does particle dynamics at the level of atoms and molecules. Including every atom and molecule in a numerical model of blood circulation is not feasible, even using today’s fastest computers. So applied mathematician George Karniadakis of Brown University and his colleagues have simplified. They begin on a slightly larger scale—bundling atoms and molecules into groups of 10 or more—and model blood circulation. Their technique, aided by scientists at Argonne National Laboratory, is producing new insight into the ways that diseases disrupt normal blood flow. Karniadakis spoke with American Scientist contributing editor Catherine Clabby about the work.
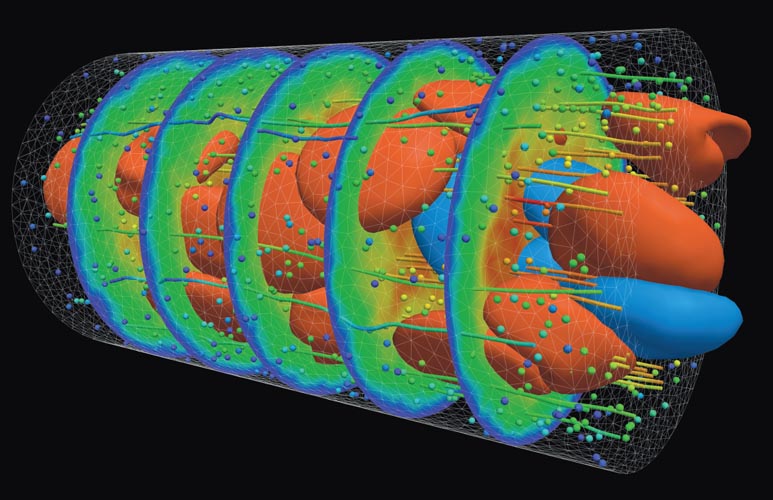
Image courtesy of Argonne National Laboratory.
What are a few of the biggest challenges to modeling blood flow through vessels?
Blood is a really complex fluid with many yet-unexplored properties. Any modeling errors greatly affect simulation results. For capillaries and vessels such as arterioles and venules, which connect capillaries to arteries and veins, the difficulty is how to scale up the molecular-based modeling. So we have developed a multiscale approach to capture the effects of both the flow and the particle dynamics and to quantify their interactions. The geometric complexity of the human arterial tree is another big challenge. For large arteries, MRI scans can be used to extract and reconstruct patient- specific arterial geometries, but this is not possible at the small scale. Finally, the viscous and elastic properties of blood cells and arterial walls introduce big uncertainties.
See a video of the work below:
What is your multiscale technique, and how could it be medically useful?
We need to span a range of spatial and temporal scales so that we can numerically portray many components accurately. That includes proteins such as spectrin, which is part of the cytoskeleton of red blood cells, and fibrinogen, which is involved in blood coagulation. At the same time we need to reach up to the vessel scale, and we need to model how those two scales influence one another. There are no standard ways of doing that. But there have been some recent advances in coarse-graining molecular dynamics methods: ways to represent systems with less than all-atom resolution. These methods do a good job of modeling blood cells, plasma, proteins, and arterial walls while simplifying the computational complexity. The coarser-grain approach still requires supercomputers to carry out realistic simulations, but it makes the problem more manageable. We have used these models to explore what specific changes to blood flow occur in vessels affected by sickle cell disease, malaria, and brain aneurysms.
What have your new simulations revealed about how diseases disrupt blood flow?
One recent example is new insight into what causes painful episodes in people with sickle cell disease. Healthy red blood cells are round and flexible, and easily change shape to move through even the smallest blood vessels. Among people with sickle cell disease, blood cells can be hard, sticky, and abnormally shaped. They resemble sickles. The common wisdom has been that this distorted shape causes blood cells to get stuck in tiny vessels and block the flow of oxygenated blood. The blockages lead to severe pain, tissue damage, and sometimes fatal organ damage.
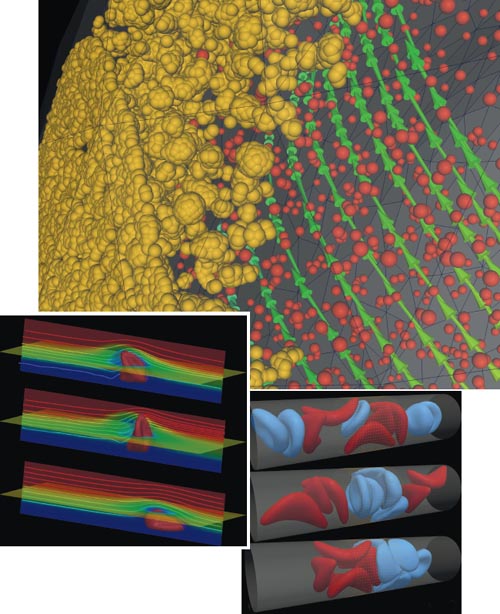
Images courtesy of Argonne National Laboratory.
With our modeling we could explore how qualities of multiple blood cells changed by sickle cell disease affect these blockages. We concluded that a subset of red blood cells plays an important role. They do not become rigid but do become stickier because of sickle cell disease. Receptors on surfaces of these cells cause them to stick to capillary walls, narrowing the diameter of some vessels. Sickle-shaped cells then stack up behind them, creating blockages. The two work in tandem, but the sticky cells start the blockage.
What about infectious diseases—can they affect blood flow as well?
We have also quantified some of the biophysical characteristics of blood cells altered by the parasite that causes malaria. We have observed that red blood cells infected by Plasmodium falciparum are as much as 50–100 times stiffer than healthy ones. With the loss of elasticity, these cells cannot pass through capillaries, so they block them. We also are making progress in understanding how blood platelets collect on the surfaces of blood vessels in the carotid arteries and in the brain during the development of cerebral aneurysms, a risk factor for strokes.
Has it been challenging to create visualizations that capture the complexity of this research?
There are no existing tools for the kind of multiscale visualizations needed to capture simultaneously details of protein dynamics and also cell or vessel dynamics. Joseph Insley, primary software development specialist, and Michael Papka, director of the Argonne Leadership Computing Facility, our collaborators at Argonne National Laboratory, are pioneering multiscale visualizations. Their work helps us observe interactions between protein, cell, and vessel dynamics that we cannot extract using standard software. Such visualizations have helped us devise new numerical methods to capture variation in blood flow.
What are your wider goals for this kind of biological modeling?
Our goal is twofold. We want to fully understand, at the molecular and arterial level, all the biophysical aspects of blood flow that are affected by the diseases we currently study. And we want to develop a general computational framework that, together with laboratory microfluidic measurements, can be used to better understand effects from other diseases. Those effects include microcirculation changes associated with the human immunodeficiency virus (HIV), diabetes, and even metastatic cancer. Our ultimate objective is to develop a patient-specific, multiscale computational framework that will aid researchers and clinicians in the prognosis of hematologic disorders.
Click "American Scientist" to access home page
American Scientist Comments and Discussion
To discuss our articles or comment on them, please share them and tag American Scientist on social media platforms. Here are links to our profiles on Twitter, Facebook, and LinkedIn.
If we re-share your post, we will moderate comments/discussion following our comments policy.