Regulatory DNA Variants in Disease: Too Much (or Too Little) of a Good Thing
By Brian J. Abraham
Decades of research into genetic disorders have scrutinized but a tiny part of the human genome—the part with the code for making proteins. This tiny part yielded the causes of sickle-cell disease and hemophilia and inspired a slew of labs to seek causes for more diseases in protein-coding DNA. But those labs’ quests returned surprising results.
March 16, 2015
Macroscope Biology Genetics
In the quest to know the causes of human disease, sometimes it’s about quantity instead of quality. Decades of research into genetic disorders have scrutinized but a tiny part of the human genome—the part with the code for making proteins. This tiny part yielded the causes of sickle-cell disease and hemophilia and inspired a slew of labs to seek causes for more diseases in protein-coding DNA. But those labs’ quests returned surprising results: Disease-causing DNA doesn’t always make a broken protein. Instead, altering the amount of protein being made from genetic instructions may cause as much trouble as altering the protein produced. Studying changes in DNA that affect protein production might hold the clues to the causes of an unprecedented number of genetic diseases.
Genetic diseases are caused by variations in the arrangement and order of DNA—the chemical set of instructions for running cells and forming an organism. The building blocks of DNA are called nucleotides, and variations in the order of these nucleotides can be either inherited from your parents, or can occur somatically to the DNA in your own cells. A random variant in DNA has roughly a four-percent chance of altering a region encoding a protein. Even if this variant occurs in protein-coding DNA, it doesn’t guarantee a change in the protein itself.
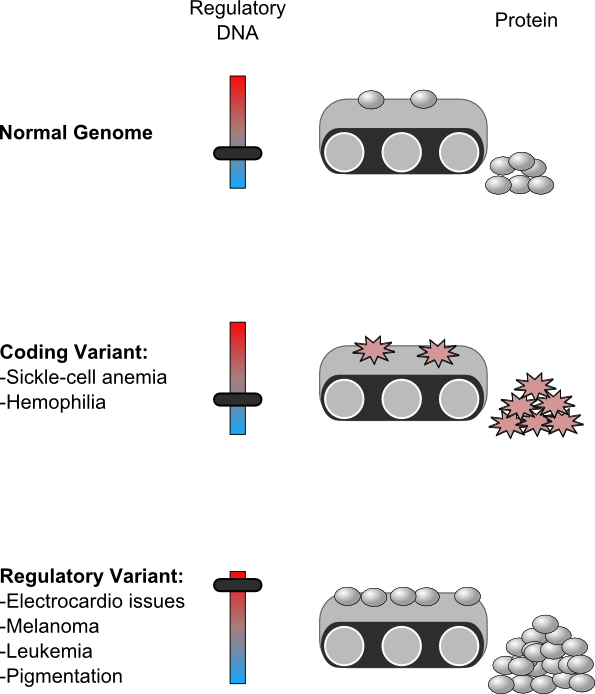
Graphic by Brian J. Abraham
Even though only a tiny fraction of your DNA encodes proteins, this fraction is the most common first place to look for a DNA variant that might cause a disease. If a protein’s code is altered by inherited variants or somatic variants, it could affect the way that protein functions. Because proteins form the parts and machines of the cell, a broken protein might leave a job undone or done wrong.
The first diseases scientists could link to DNA variants were those where protein structure and function were altered. Between the 1940s and 1960s, researchers found that sickle-cell disease is caused by changes to the DNA encoding hemoglobin, and hemophilia, is caused by changes to the DNA encoding coagulation proteins. So, the quest for more links between disease and DNA began, and the first results were surprises. Inventors developed technologies to compare the DNA of groups of people with a disease to the DNA of groups of people without that disease. When researchers mapped where the DNA variation between patients versus controls fell, they found an enormous majority of these variants were not actually in protein-coding DNA.
Some parts of noncoding DNA—the other 96 percent of the genome—have other jobs; this includes regulatory DNA, which controls when a protein is made from a gene’s instructions. Regulatory DNA binds protein signals called transcription factors, and tells the nucleus’ machinery to produce a copy of a specific protein. Recent papers (published in Science Magazine, Cell, and Nature) including those just released from the Epigenome Roadmap project ("Integrative analysis of 111 reference human epigenomes" and "Genetic and epigenetic fine mapping of causal autoimmune disease variants"), say that variants linked to many diseases tend to fall in this regulatory DNA.
Scientists have proposed how these variants in regulatory DNA might be causing problems. Regulatory DNA can entice specific transcription factor proteins to bind them if they contain certain short sequences of nucleotides—DNA sequence motifs. These sequences are usually between 4 and 12 nucleotides long. Changing just one nucleotide can cause or prevent a transcription factor from binding. If DNA variants exist in regulatory DNA, some of them might alter transcription factor-binding motifs, change which proteins bind, and alter protein production levels. If a cell requires a certain amount of a given protein, changing how or how frequently it is produced can be catastrophic.
Several recent cases of variants in regulatory DNA imply they can contribute to disease.
The heart’s contractions are controlled in part by electrically charged ions flowing through protein gates on the surfaces of cells, and altering these gates can prove costly (example 1 and example 2). Binding of transcription factor proteins at regulatory DNA controls production from genes encoding ion gate proteins SCN5A and SCN10A. A variant in regulatory DNA seen in patients with electrical conduction issues in their hearts alters the ability of this regulatory DNA to drive production. Perhaps these variants alter production of these protein gates and affect heartbeats.
Cancer is perhaps the quintessential disease of broken DNA. So, it seems logical that DNA variants in regulatory regions could be important for cancer cell survival, and, as the spotlight is beginning to shine on regulatory DNA, we’re beginning to find them.
Telomeres are caps on the ends of DNA chromosomes that correlate with a cell’s age; when telomeres get too short, the cell is pushed toward suicide. Cells with telomeres that are too long avoid this natural cell death and can progress toward cancer. The TERT protein helps control telomere length. In several types of cancers, including melanomas and gliomas (Science on the mutations in human melanoma, Science on familial and spradic melanoma, and PNAS on gliomas), a somatically acquired variant creates a binding site for a protein called ETS1 in regulatory DNA. ETS1 binds this region and causes an increase in TERT production, and presumably an increase in telomere length. Longer telomeres can put off cell death, making some cells becoming problematically long-lived.
Another research group showed that creation of a binding site for the MYB protein in leukemia cell DNA can regulate a regulator of many important genes. This variant causes increased production of the transcription factor protein TAL1, which itself regulates production from a large number of genes and keeps these leukemia cells in an "undifferentiated" state. Since they’re stuck in immaturity, they do little more than accumulate. Interestingly, these same cells that rely on TAL1 production are sensitive to experimental drugs that target the machines that do the producing, suggesting a promising potential avenue for treating some leukemias.
Even non-disease traits like hair and skin pigmentation may be controlled by variants in regulatory DNA. Mice with DNA containing one version of a regulatory region that controls production of a signaling protein, KITLG, have significantly darker hair color than mice with a single DNA nucleotide difference. This single nucleotide creates or destroys a sequence motif bound by LEF1 protein and might explain some genetics of blonde hair in Europeans.
For a long time, it has been easier for scientists to examine protein-coding DNA, but technology has enabled us to look with the same scrutiny at noncoding DNA, and the results are tantalizing. It’s possible the causes to many diseases are hiding in the shadows of the genome, causing differences in protein quantity instead of quality, all waiting to be hit by science’s limelight.
American Scientist Comments and Discussion
To discuss our articles or comment on them, please share them and tag American Scientist on social media platforms. Here are links to our profiles on Twitter, Facebook, and LinkedIn.
If we re-share your post, we will moderate comments/discussion following our comments policy.