Science Needs More Moneyball
By Frederick M. Cohan
Baseball’s data-mining methods are starting a similar revolution in research
Baseball’s data-mining methods are starting a similar revolution in research

DOI: 10.1511/2012.96.182
The Moneyball story, in book and film, champions a data-mining revolution that changed professional baseball. On the surface, Moneyball is about Billy Beane, the general manager of the Oakland A’s, who found a way to lead his cash-strapped club to success against teams with much bigger payrolls. Beane used data to challenge what everyone else managing baseball “knew” to be true from intuition, experience and training. He pioneered methods to identify outstanding players he could afford because they were undervalued by the traditional statistics used by the baseball elite.
This film was marketed as a sports movie. When I saw it, I knew right away what Moneyball is really about: the thrill and triumph of data mining. It’s an instructive tale of how existing data can be examined for meaning in ways that were never intended or imagined when they were originally collected. Beane and his colleagues challenged the time-honored trinity of batting average, home runs and runs batted in (RBIs) as the essence of the offensive value of a player, replacing these statistics with newer measures based on the same data. They worked off theories developed by baseball writer and historian Bill James, who posited in the 1970s that the traditional stats were really imperfect measurements. James’s approach didn’t just replace one intuition with another. He let the game decide which stats did the best job of predicting offensive output.
This approach is not easy. Trying to directly predict the number of games won would confound the skill of a team’s offense with its pitching and fielding. James figured that one could test each offensive stat by trying to predict the total number of runs produced by each team over the course of a season, thus eliminating any effects of defense. It turned out that on-base percentage and slugging percentage were far superior to any individual offensive statistics used up to that point. James and others similarly devised statistics for pitching and fielding that were more independent of context.
Beane’s use of the new statistics is appealing because it defeated the wisdom and training of other industry experts. His approach is summed up in one of the best scenes from the Moneyball film. Armed with his new data-mining methods, Beane challenges other talent evaluators about a player they all deem “good.” A scout counters him, praising the player’s swing. Beane’s reply: “If he’s such a good hitter, why doesn’t he hit good?”
In other words, expert intuition aside, the data don’t lie.
As I see it, the baseball revolution produced an “idiot’s guide” to creating a team roster—a handbook based on things one can learn not through decades of experience and intuition but by applying general quantitative methods. It’s the same kind of approach we should employ more in the sciences. Mountains of data and a capacity for analyzing them have also become available to science in the past few years. Data are now poised to trump the intuition of experts and the “facts” that scientists have championed over the years.
For instance, consider my own field, biology. Every biologist “knows” what a species is—a group of organisms that can successfully produce viable and fertile offspring. Biologists have long believed that species defined this way represent the fundamental units of ecology and evolution.
In the case of evolutionary microbiology (my specialty), it is particularly important to be able to recognize all the fundamental units of ecology among closely related bacteria. We especially need to distinguish those that are dangerous from those that are not and those that are helpful from those that are not. Indeed, we would like to identify all the bacterial populations that play distinct ecological roles in their communities.
As in baseball, the discovery of bacterial diversity has experienced a transition from relying on the subjective judgment of experts to objective and universal statistical methods. Originally, discovery and demarcation of bacterial species required a lot of expertise with a particular group of organisms, involving difficult measures of metabolic and chemical differences. To make the taxonomy more accessible, decades ago the field complemented this arduous approach with a kind of idiot’s guide, where anyone could use widely available molecular techniques to identify species—for example, a certain level of overall DNA sequence similarity.
One popular universal criterion (among others) is to identify species as groups of organisms that are at least 99 percent similar in a particular universal gene. The problem is that—like the case of baseball where batting average, RBIs and home runs were used to supplement expert knowledge—nobody in microbiology tested whether the new molecular techniques actually came closer to solving the problem of recognizing the most closely related species.
Unfortunately, microbiology’s current DNA-based idiot’s guide, as well as the expert-driven metabolic criteria that preceded it, has yielded species with unhelpfully broad dimensions. For example, Escherichia coli contains strains that live in our guts peaceably, as well as various pathogens that attack the gut lining and others that attack the urinary tract. Moreover, established fecal-contamination detection kits that are designed to identify E. coli in the environment are now known to register a positive result with E. coli relatives that normally spend their lives in freshwater ponds, with little capacity for harming humans. And E. coli is not alone—there is a Yugoslavia of diversity within the typical recognized species: Much like the veneer of a unified country that hid a great diversity of ethnicities and religions, E. coli (and most recognized species) contains an enormous level of ecological and genomic diversity obscured under the banner of a single species name.
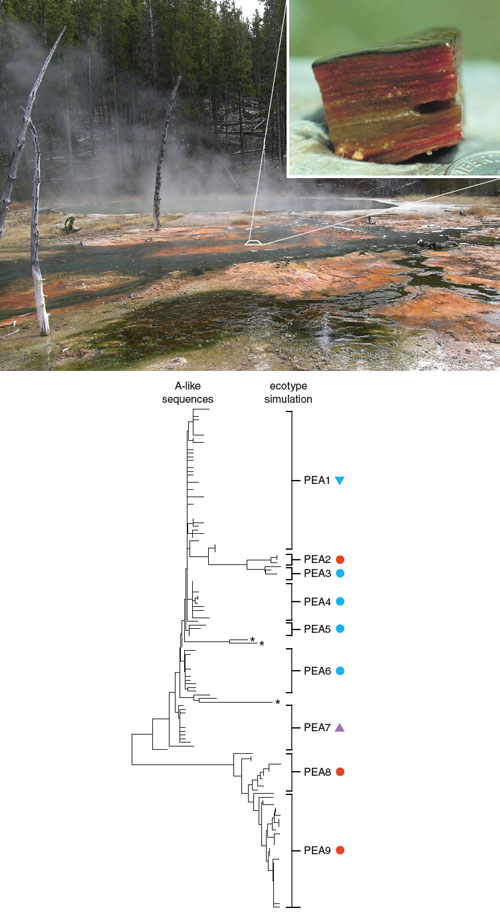
Photographs courtesy of David Ward.
We can fix this confusion the same way that baseball improved its data analysis: by letting the game—or in our case, nature—decide which stats best predict what we most want to know. In microbiology the trick is to let the bacteria tell us what DNA sequence approach most accurately identifies the bacteria that are significantly different in their habitats and ways of making a living. Two teams, including Martin Polz’s group at the Massachusetts Institute of Technology and my group at Wesleyan and Montana State Universities, have developed computer algorithms for identifying groups of bacteria specialized to different habitat types within an officially recognized species. These algorithms reject the expert-based criteria for how much diversity should be placed within a species. Instead, they analyze the dynamics of bacterial evolution to let the organisms themselves tell us the DNA sequence criterion that best demarcates ecologically distinct populations for a particular group of bacteria.
Another opportunity for discovery in biology through data mining stems from the new Human Microbiome Project. Here, DNA sequences are collected from various bacteria-laden human habitats, such as the gut, mouth, skin and genitals, with samples taken from individuals of different age, sex, health, weight and diet.
For example, Dusko Ehrlich of the French National Institute for Agricultural Research and his colleagues recently analyzed the bacterial genes purified from the feces of 39 humans from six European countries, amounting to about 100 million bases of bacterial DNA per person. They attempted to identify bacterial biochemical functions associated with age and body mass. Their intuition suggested various guesses for the identity of these genes, which were largely supported, but data-driven methods identified genes that gave much stronger relationships. One important data-driven discovery indicated a negative relation between obesity and the microbes’ capacity for harvesting energy.
Ongoing massive sequencing projects in human, marine and soil environments allow us to characterize the diversification of bacteria: to discover the most newly divergent bacterial species, to characterize them as specialized to different habitats and to identify the biochemical functions most important in each habitat. However, the approach depends critically on how well we describe the habitats we sample.
Beyond the field of microbiology, data- mining revolutions are extending across the natural and social sciences (although meteorology and economics, with decades-long access to mountains of data, are still the granddaddies of this approach). In the social sciences, it is particularly interesting to see how data mining has recently helped linguists analyze how words are actually used in writing and speech—for example, as seen in the challenge of producing a dictionary. Traditionally, analysis of language use has involved assessment of written texts, usually from a canon of books accepted by experts as exemplars of “proper” usage, a step that required an army of volunteers who sent in quotations to the dictionary editors. Then the appointed set of language experts made subjective decisions about new usage—what is acceptable, what is vulgar and what is vile. A data revolution in linguistics is freeing us from needing the army of volunteers, as well as from the opinions of the learned experts. Language analysis is heading toward a data-driven idiot’s guide that can decide on acceptable usage based on what is actually accepted in writing and in speech.
Various corpora of written and spoken language have emerged online, and these allow extensive analysis of how and where words are used. Entire uploaded texts can be searched and analyzed. The largest is the Oxford Corpus, launched in 2006 and covering texts from the entire Anglosphere. The U.S.-centered Corpus of Contemporary American English (COCA) features a user-friendly website ( http://corpus.byu.edu/coca/). These corpora, when searched, give a 10-word neighborhood around each use of the word, which yields much information. For instance, a searcher can see whether the word is used in the singular or plural form, as well as words that are frequently co- located with it and so on. In Damp Squid, Jeremy Butterfield describes how these corpora can yield a picture of English (or potentially any language) as it is actually used, as validated by the entire community of writers and speakers.
One way that corpus-based analysis bucks expert opinion is in deciding when an evolutionary change in usage has become acceptable simply by the criterion of being frequently accepted. For example, the word “criteria,” on entering the English language from Greek, maintained its original meaning as the plural of “criterion.” Cringe though we may, our own experiences plus analysis of the Oxford Corpus show that use of “criteria” as the singular is catching up on its use as the plural. The corpus also allows us to note changes in old expressions that still hold meaning for us, but only if we change the words a little. Shakespeare’s “in one fell swoop” is still a popular phrase four centuries later, but only through changing the obsolete adjective “fell” to one that sounds similar and holds a similar meaning, which is “foul.” Despite the resistance of experts, the language is de facto evolving, and the corpus allows us to validate these changes.
As useful as the idiot’s guide approach has been across fields, gleaning meaning from old data serves up severe challenges. Difficulties can arise because at the time events happened, the data recorders did not anticipate that the information would be analyzed in ways not yet imagined. In cases stretching across baseball, biology and language, important items were not reported or, in some cases, observed at all. There is a twin problem to using past data, which is a communitarian challenge—appreciating that data are often used in ways unimagined at the time of collection, how can we make the data we record today more usable and valuable in the future?
Image courtesy of the author.
As baseball and the sciences have taken an interest in mining old data for new insights, it has turned out that the old data sets are often sufficiently complete for us to discover new “laws” of baseball or science. Yet in far too many cases, fresh scrutiny of old data reveals painful omissions proving that science has missed an opportunity.
In retrospect, I am amazed at how little interest baseball and biology have shown for the future use of data. In baseball, the traditional play-by-play record of games was all that was reliably available until 1988, when the pitch-by-pitch record became the standard. The new record turned out to be important in many ways—for example, in managing a pitcher’s productivity, health and longevity.
Until recently, biology was equally shortsighted in its data collection; this has created a problem for biologists who would like to analyze other scientists’ published data. For example, Cathy Lozupone and Rob Knight at the University of Colorado figured out from analyses of others’ data that the most difficult evolutionary transition in the history of bacteria has been from saline to non- saline environments and vice versa. However, because the original researchers did not record the actual salinity levels, Lozupone and Knight could not pinpoint the precise concentration of salinity that has been most difficult to cross.
Previous standards of data collection in biology were typically limited to what might be interesting for the experiment at hand or perhaps for some future experiment in the same lab. Today, biologists are increasingly expected to anticipate likely uses by others of the data we gather and are taking pains to do so, but this forethought is not easy.
I recently met with Hilmar Lapp, a database expert at the National Evolutionary Synthesis Center (NESCent), and discussed how researchers could avoid omitting important elements of data. He said that it is too much to expect, in the case of biology, for one researcher to think to include all the observations worthy of recording for posterity; he suggests what is needed is a “crowd intelligence.” Accordingly, NESCent and other organizations have sponsored working groups to pool ideas and propose standards and directions of biological data collection in novel areas of inquiry—that is, to foster crowd intelligence. For example, the Genome Sequencing Consortium recently established standards for recording environmental data when genes and genomes are sampled; earlier action might have avoided the debacle of the missing salinity data Lozupone and Knight encountered.
In some cases, we do not have data on old events, not because of a lack of imagination but because the appropriate technology was not available at the time. In the case of baseball, the new, high-tech Advanced Value Metrics (AVM) system automatically describes each hit ball by its trajectory, velocity and point of hitting the ground. The AVM description of a hit allows analysis of how frequently a fielder can catch a ball that usually ends up being a double. But no one could analyze the skill of fielders at this level prior to the advent of this technology.
Until recently in biology, a lack of microbiological technology limited plant ecologists’ understanding of the factors allowing a particular plant species to grow. Plant ecologists discovered only recently that the success of many plant species in nature is determined by helpful and harmful microbes that live in the soil. Therefore, decades of studies trying to understand the successes and failures of plants came up short because they failed to collect data on soil microbes.
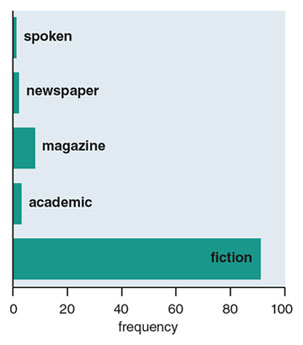
Illustration by Barbara Aulicino..
In linguistics, the lack of technology for audio recording has hindered an analysis of spoken English usage over time. You might think that dialog written in novels and stories would be a good substitute for actual sound recordings; these pages are frequently as good a record as we will get. However, it is discouraging that a corpus-based analysis of word usage in speech versus fiction by lexicographer and author Orin Hargraves has shown that certain clichéd phrases, which appear to mimic spoken language, are actually used far more frequently in literature than in real life. For example, hardly anyone really says “he bolted upright” or “she drew her breath,” but these forms are found with surprisingly high frequency in literature. Consequently, an unbiased, corpus-based account of spoken English usage begins with abundant voice recording in the 20th century.
Analyses of huge data sets allow us to move beyond our previous understanding, which was based on much less data than we have available to us today. There is so much possibility for a data-driven explosion of understanding of games, creatures and words by explorers today and in the future. We owe these future explorers the best and most complete record of life today that we can offer.
The Moneyball film opens with wisdom from Mickey Mantle: “It’s unbelievable how much you don’t know about the game you’ve been playing all your life.” Surely the same is true for many in the natural and social sciences, pondering the areas they have been studying all their careers.
Click "American Scientist" to access home page
American Scientist Comments and Discussion
To discuss our articles or comment on them, please share them and tag American Scientist on social media platforms. Here are links to our profiles on Twitter, Facebook, and LinkedIn.
If we re-share your post, we will moderate comments/discussion following our comments policy.