Cell by Cell, Life Appears
By Catherine Clabby
Thin sheets of laser light illuminate the incredibly complex process of embryonic development.
Thin sheets of laser light illuminate the incredibly complex process of embryonic development.

DOI: 10.1511/2014.111.466
In the broadest sense, it is obvious how embryos develop. A single cell cleaves into multiple cells, which subdivide into daughter cells. The pattern repeats again and again, giving rise to specialized cell lines and ultimately to the systems that sustain each organism. Things get murky, however, if you try to track that transformation cell by cell. Complex changes occur on scales of submicrometers in just hundreds of milliseconds, pushing the limits of scientific observation.
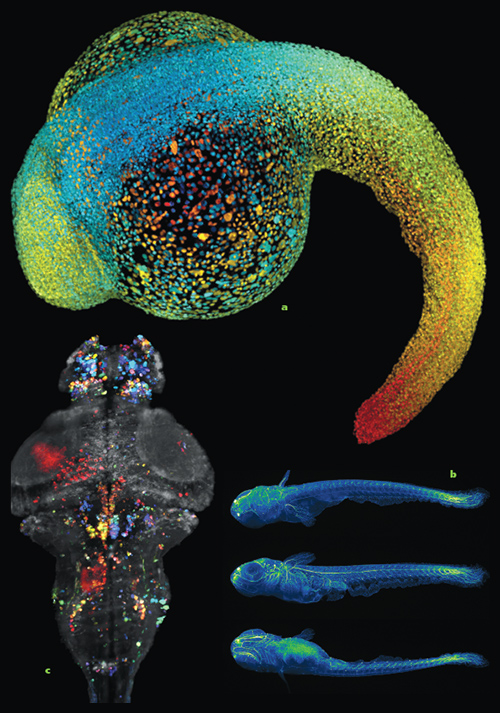
Images courtesy of Phillip Keller, Howard Hughes Medical Institute.
Philipp Keller, a biophysicist at Howard Hughes Medical Institute (HHMI), is pushing back. He and his colleagues are combining a technique called light-sheet microscopy with novel computational strategies to record early cell division, cell migration, and the emergence of early structures in embryos. The development of nervous systems particularly intrigues him. “Among all the tissues and systems in all early animals, the nervous system is especially fascinating in its complexity and its function. It coordinates everything,” says Keller, who runs a lab at HHMI’s Janelia Farm Research Campus in Ashburn, Virginia.
Light-sheet microscopy illuminates and records one very narrow sliver of a specimen at a time. So-called sheets of laser light, measuring just a few micrometers wide, strike one slice at a time from multiple planes; specialized detectors capture and record images of each slice. Those images can be combined to assemble a complete digital rendering.
In 2004 a research team led by Ernst Stelzer at the European Molecular Biology Laboratory demonstrated the technology’s promise in developmental biology. The mix of limited exposure to damaging light and good-resolution detail is especially useful for imaging living embryos. Early studies captured the beating heart of an embryonic Japanese rice fish, the medaka, and structures inside in the fruit fly embryo, which is more opaque.
While he was a doctoral student in Stelzer’s lab, Keller refined the technology. He sped up imaging and produced sheets of light with more uniform intensity, which improved the image quality. He also modulated the amplitude of the lasers to remove the scattered light in images and provide high-contrast views deeper within a sample. In 2008, Keller published a step-by-step image sequence showing the 24-hour transformation of 32 cells into more than 20,000 cells in a zebrafish embryo, a widely used model organism in developmental biology.

Images courtesy of Phillip Keller, Howard Hughes Medical Institute.
Since then, Keller and his collaborators have recorded nervous system development in zebra- fish embryos and have digitally reconstructed full fruit fly embryos (they can be viewed online: http://www.janelia.org/digitalembryo). In 2013, Keller and his HHMI colleague Misha Ahrens recorded the activation of neurons across a larval zebrafish brain, the first time such single-cell scale activity was captured in a vertebrate. And just this year Keller’s HHMI team developed open-source software that both tracks all types of cells in developing embryos and can isolate specific cell lines. They used it to trace the development of nearly 300 neuroblasts, precursors to neurons, in fruit fly embryos.
Keller next wants to integrate light-sheet imaging with gene expression studies, using fluorescing protein tags to capture genetic activity in individual cell nuclei during embryonic development. Observing where and when regulatory genes switch on could reveal processes essential to the differentiation of a few cells into a complex organism. “In physics we have a few rules that can explain a lot of observable phenomena. In biology there could be similarly fundamental principles of development,” Keller says.
Watch a short video of the very early development of a Drosophila embryo:
Video courtesy of Kristin Branson, Fernando Amat, Bill Lemon, and Philipp Keller at the Howard Hughes Medical Institute / Janelia Research Campus
This time-lapse recording captures development of a Drosophila embryo from the blastoderm stage to about five hours after eggs holding embryos were laid. Green spheres superimposed on the image note the location, movements, and divisions of precursors to neural cells in a developing nervous system. Light-sheet microscopy, computer-aided cell tracking, and the tagging of cell nuclei with fluorescent proteins are some of the tools that make the observation possible.
Click "American Scientist" to access home page
American Scientist Comments and Discussion
To discuss our articles or comment on them, please share them and tag American Scientist on social media platforms. Here are links to our profiles on Twitter, Facebook, and LinkedIn.
If we re-share your post, we will moderate comments/discussion following our comments policy.