See How They Grow
By Catherine Clabby
Nearly nondistorting fluorescent tags can capture growth patterns in bacterial cell walls.
Nearly nondistorting fluorescent tags can capture growth patterns in bacterial cell walls.

DOI: 10.1511/2013.102.226
Fascination with bacteria is on the rise for good reason. Thanks to fast and more affordable DNA sequencing, it’s now clear that bacteria are more varied, more abundant and more influential on Earth than anyone knew. One estimate says five million trillion trillion of the single-cell prokaryotes dwell here. Some of the microbes are helpful to other organisms; others are harmful. Now Indiana University scientists, with collaborators in Spain, have put the one-cell life forms in better focus. Using nearly nondistorting fluorescent tags, they can capture growth patterns in bacterial cell walls. The advance could help scientists improve antibiotic drugs, develop new insights into microbe development and better observe which bacteria thrive in a given environment. In a written exchange, chemist Michael VanNieuwenhze, microbiologist Yves Brun and doctoral biochemistry student Erkin Kuru at Indiana explained the new technique to American Scientist contributing editor Catherine Clabby.
How does your method improve on previous efforts?
In order to study the mechanisms of bacterial growth, researchers had developed methods to label sites of active synthesis of peptidoglycan. This is a polymer in bacterial cell walls that defines cell size and shape and provides it with mechanical strength to resist cell envelope breakdown. Previous labeling methods relied on fluorescently modified antibiotics or modified cell wall precursors that altered the very process they were meant to monitor and often required probe concentrations that were toxic to cells. Our method uses fluorescent dyes bonded to compounds called D-amino acids (FDAAs) as probes. The probes are incorporated by enzymes involved in the peptidoglycan biosynthetic pathway. This process is not toxic to cells and can therefore be used to monitor cell wall growth in live cells without altering the process itself.
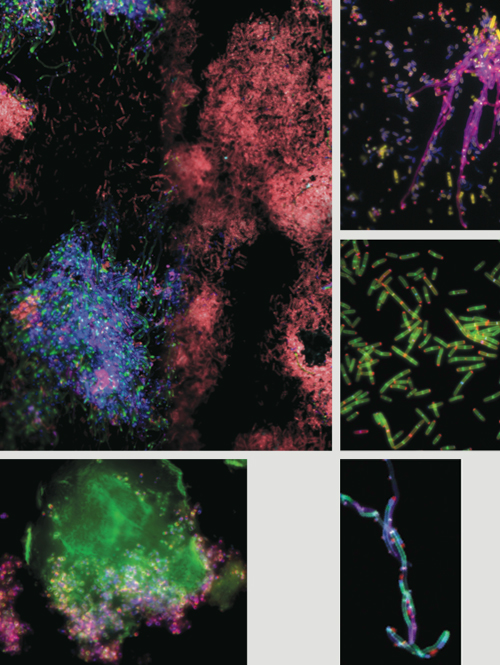
Images courtesy of Michael VanNieuwenhze, Yves Brun and Erkin Kuru.
The spatial organization and coordination of peptidoglycan synthesis within bacterial cells is important for optimal growth and to prevent cell envelope breakdown. The colors enable real-time tracking of the timing and location of polymer synthesis. The new technique allows us to monitor how peptidoglycan synthesis responds to antibiotics and environmental changes. It also provides previously unavailable information regarding modes of bacterial growth.
Could this technique deliver new insights into bacterial development?
Bacteria exhibit myriad cell shapes. Some look like stars, flat squares or telephone cords; others have antenna-like extensions of the cell envelope. It is clear that these shapes provide selective environmental advantages because they are faithfully reproduced, but the function of the vast majority of shapes and how they are generated remains a mystery. We have had only a rudimentary understanding of the mechanisms used by bacteria to make the simplest shapes: spheres and short rods. We lacked a reliable method to determine what part of a cell grows at what time to generate more complex shapes. In addition, modes of growth have been observed only in a few model systems due to the limitations of the earlier methods. FDAA labeling enables us to rapidly study growth modes of multiple bacterial species. With that and with the power of comparative biology, we can explore basic growth mechanisms.
What is the promise here for improving antibiotics?
Antibiotics that target the peptidoglycan biosynthetic machinery have been the most widely used and effective class of antibiotics to date. This is partly due to the fact that peptidoglycan biosynthesis is unique to bacteria. An agent that inhibits any step in this pathway is toxic only to bacterial cells. Our method accelerates the study of the mechanics of peptidoglycan biosynthesis. A better understanding of this process can lead to discoveries of yet unknown vulnerabilities in bacterial growth mechanisms. That may reveal new killing mechanisms and effective natural antibiotics. It may also help develop new antibiotics to exploit the vulnerabilities. In fact, our work on the mechanisms of FDAA incorporation suggests that FDAAs are specifically directed into the active site of essential cell wall enzymes. Thus, FDAAs may provide a template for the design of D-amino acid antibacterial agents.
What are the merits to seeing which bacteria are active and inactive in the same environment?
Bacteria are everywhere. But depending on their growth requirements and variation in the supply of nutrients and other conditions, some bacteria will be dormant and others will be active in the same environment. Knowing which populations are and are not growing provides an assessment of environment quality. Furthermore, developing methods to improve the growth of beneficial species of bacteria and to impede the growth of detrimental species is important.
When will this method be available to other scientists?
A provisional patent application has been filed and the full application will be filed in the near future. A commercial supplier for the probes remains to be identified. However, we have been supplying FDAAs to other researchers on a case-by-case basis to enable the immediate use of this powerful new tool.
Click "American Scientist" to access home page
American Scientist Comments and Discussion
To discuss our articles or comment on them, please share them and tag American Scientist on social media platforms. Here are links to our profiles on Twitter, Facebook, and LinkedIn.
If we re-share your post, we will moderate comments/discussion following our comments policy.