Replaying Evolution
By Zachary D. Blount
Is the living world more a result of happenstance or repeatable processes?
Is the living world more a result of happenstance or repeatable processes?

Amazon’s television series The Man in the High Castle, based on the classic novel by Philip K. Dick, presents a nightmarish alternative 1962 in which the triumphant Nazi and Japanese empires occupy a fractured, defeated United States.
This alternate history is spun from the imagined consequences of a minor change in a real event. On February 15, 1933, Giuseppe Zangara opened fire on president-elect Franklin D. Roosevelt in Miami, Florida. Zangara was only 25 feet away, but his attempt failed because he shared the wobbly bench on which he stood with a woman who, as she strained to see, jostled the bench at just the right time to spoil his aim. The show’s version of history did not include the fortuitous jostle. Although the result of such a change might not have been the dystopia the show envisions, history would have been quite different had Roosevelt died that day.
Human history has been wrought from the particulars of unique events and personalities. Indeed, the historical record is rife with instances like the attempt on Roosevelt’s life, where even slight changes could have dramatically altered the course of events. These instances illustrate how the existence of the current world depended on the process of history, linking past to present in a complex web of causality. In other words, human history is contingent. Contingency, philosopher John Beatty has written, essentially means that history matters “when a particular future depends on a particular past that was not bound to happen, but did.” It arises because the future flows causally from the past, but many futures are possible at any given time, and which one comes to pass is determined by the precise, chance-laden way in which a complex tangle of improbable events interacting in improbable ways plays out. Contingency is why we can more or less explain the past, but the future is unpredictable.
Even a cursory survey makes it hard to deny that contingency has played an important role in human history. But human history is embedded within the 4-billion-year evolutionary history of the living world. Has this grander history of life been similarly subject to contingency, making the modern living world as a whole as much a unique product of coincidence and accident as the current state of humanity? It’s a startling question with mind-boggling implications. We tend to think of natural phenomena as regular and deterministic, proceeding from a beginning along an inevitable path, sure as the planets in their orbits or a ball rolling down a hill. Whether or not the process by which life has developed and evolved on Earth is really so deterministic, however, is an open question. Indeed, whether evolution itself is contingent in the same way as human history is one of the most vibrant and important debates in biology.
Evolution is different from many other natural phenomena in that it is fundamentally historical. Like human history, evolution plays out over time and involves a fundamental tension between chance and necessity. Natural selection deterministically adapts organisms to their environment by incessantly winnowing the wheat of beneficial variation from the chaff of the neutral or the detrimental. The variation on which natural selection works arises from random genetic changes, however, and even the most beneficial mutations can be lost, especially in small populations, due to the lottery-like effects of genetic drift—random fluctuations in gene frequencies. Moreover, just as in human history, evolution must take place in the context of what has evolved before, which can alter the prospects for future change. Finally, evolution is a process that is strongly affected by changes in the world within which it occurs. Evolution has taken place within the broader history of Earth, which has included events such as asteroid strikes and capricious, even cataclysmic, environmental change.
Evolution’s chancy, historical nature suggests that contingency plays an inevitable role. The debate in biology has been about how large that role is. This debate has been framed using a thought experiment, called “replaying life’s tape,” proposed by the late Harvard paleontologist Stephen Jay Gould in his 1989 book, Wonderful Life. Imagine one could go back in time to the distant past, the thought experiment goes, and then let evolution run its course again. What would happen? There are two basic options, depending on the role one assigns to contingency: Either the living world we know reevolves, or something else arises. Gould argued that evolutionary outcomes are highly sensitive to the details of history, and so contingency is extremely important. Therefore, he believed, each replay would result in a different living world because all the chance factors involved would make it unlikely for the same history to recur.
Illustration by Tom Dunne.
Others have suggested that the outcomes of rerunning evolution would be far more striking in their similarities than their differences. They have pointed out that evolution is categorically different from human history because of the strong, deterministic power of natural selection. Cambridge paleontologist Simon Conway Morris and others have argued that natural selection will find the same biological solutions over and over again regardless of variations in history. As evidence, they point to the remarkable number of instances in nature in which evolution has independently converged on the same traits and adaptations. Moreover, Conway Morris has speculated, convergence might also indicate that the range of viable evolutionary outcomes is constrained, so that “the evolutionary routes are many, but the destinations are limited.”
Gould’s thought experiment highlights why it is so important to understand the role of contingency in evolution. Contingency reduces evolutionary repeatability, so it has far-reaching implications for what sort of phenomenon evolution is. If evolution is highly contingent, then it is inherently unrepeatable. Even under the same conditions, evolution would end up yielding different outcomes. Although we would be able to understand an evolutionary outcome by reconstructing sequences of events as we do in human history, we couldn’t predict evolution ahead of time. If, on the other hand, contingency’s effects are negligible, then evolution is repeatable and will always converge on more or less the same outcome under the same conditions. That repeatability would make it possible to one day precisely predict evolutionary outcomes from starting points by using equations and mathematical models just as we can with phenomena in physics or chemistry. We could perhaps even predict what life-forms we might encounter on other worlds once we know what conditions are like. Of course, it is far more likely that contingency plays an intermediate role, with evolution leading to outcomes that are repeatable and convergent in some aspects, but divergent in others. The question then is, what is the balance? What biological features are likely to pop up again as predictable outcomes of evolution, and which will vary due to contingency?
Unlike historians, whose lack of time machines hampers their ability to test hypotheses about contingency’s effects, biologists have tools with which to empirically examine its role in evolution. Indeed, biologists have been investigating evolutionary contingency since the debate over it began. Much of this work has used approaches inspired by Gould’s “replay life’s tape” thought experiment to examine contingency arising within the evolutionary process itself. One avenue of research has taken advantage of how nature itself occasionally replays the tape on a small scale when similar situations emerge in separate locations. Another has exploited advances that permit ancient proteins to be resurrected, their evolutionary histories reconstructed, and the role of contingency in that evolution to be parsed out. Finally, fast-growing microbes have been used to play new evolutionary tapes in the laboratory—for example, in the Long-Term Evolution Experiment (LTEE) with Escherichia coli, on which I work. These studies have shown that although contingency is inherent to evolution, its scope and impact are variable. Indeed, sensitivity to history may itself be historically contingent and partly tied to how past evolution affects the potential for future change.
One way to study evolutionary contingency is to look for natural cases of evolution playing out under similar conditions. Archipelagos, for instance, can have dozens of islands with nearly identical environments and histories, so that the influences on natural selection are similar but evolution plays out independently on each. Over the past 20 years, Jonathan Losos of Harvard University and his colleagues have done research on the islands of the Caribbean that suggests that the effect of evolutionary contingency is more constrained than Gould had supposed.
Islands either rise from the sea as bare rock, devoid of plants or animals, or become isolated from the mainland by sea-level increase or continental drift. In the former case, an island’s biodiversity evolves as it is colonized by organisms from other islands or a mainland, and their descendants evolve to fill an island’s empty niches. In the latter case, there is much the same dynamic, albeit with preexisting inhabitants and fewer empty niches. Were the evolution of biodiversity highly contingent, then one would expect each island to evolve radically different assemblages of species, even from the same colonists. The work of Losos and his team, however, with a prominent (not to mention cute) part of the Caribbean islands’ biodiversity—Anolis lizards or anoles—shows that this isn’t the case.

Images of twig anoles courtesy of Jonathan B. Losos, with the exception of A. valencienni, which is courtesy of Kevin de Quieroz. Other images from Wikimedia Commons. Illustration by Tom Dunne.
Anoles are small, insect-eating lizards found across the Caribbean. Although the Greater Antillean anoles, which are found on the largest of the islands, are diverse, most species can be sorted into a small number of categories called ecomorphs, depending on the habitats they occupy—the ground, treetops, twigs, grasses, and so on. The lizards of each ecomorph share an array of adaptations to their respective niches. Those that run along the ground, for instance, have long legs and smaller toe pads, whereas those that spend most of their time on twigs have short limbs and tails. The lizards that belong to each ecomorph are strikingly similar across the different islands.
This pattern of similarity might have emerged because the ecomorphs evolved only once and then spread across the islands. In 1998, however, Losos and his colleagues showed that the anoles did not evolve this way. Rather, they found that the lizards belonging to a given ecomorph on one island are almost never closely related to lizards of the same ecomorph on other islands. More commonly, the closest relatives of a given ecomorph are the lizards of other ecomorphs on the same island. This pattern suggests that the assemblages of similar ecomorphs evolved independently and repeatedly on each island as colonist species diversified to fill the same open niches. The central lesson of Losos’s work is that adaptation to similar ecological conditions can lead to convergent evolution of certain traits and even communities, no matter the historical differences. But is such convergence always the case?
The Caribbean anoles have shown us much about the repeatability of evolution, but they do not fully belie the role of contingency in evolution. After all, we lack a precise, full record of their evolutionary history, which would be necessary to evaluate how that history has mattered to the patterns that Losos and his colleagues found. Moreover, we can’t manipulate the anoles’ history to experimentally test historical hypotheses. For more than a decade now, first as a graduate student and now as a postdoctoral researcher, I’ve been examining evolutionary contingency using an approach that doesn’t have these shortcomings. The broad outlines of this approach make it sound like something out of science fiction: Imagine being able to start multiple worlds anew, each identical, seeding them with a single organism, and letting evolution play out in front of us. Imagine one could save slices of the evolutionary history of each world, frozen in time, and suspended as though in amber. Evolutionary history would be much easier to reconstruct in detail. Now imagine that the organisms in those slices remain alive, allowing us, for example, to do the equivalent of directly comparing a modern human with a Neanderthal or an Australopithecus. Such direct comparisons could reveal much about how evolution proceeded, how innovations arose, and perhaps even how history takes one path rather than another.
As fantastic as it might seem, lab experiments using fast-evolving microbes make this scenario a reality. Although simplified and artificial, these experiments benefit from the rigor and control of laboratory science, making it possible to investigate questions that are difficult, if not impossible, to answer in nature. Indeed, researchers have been using these experiments to replay the tape of life, albeit on a smaller scale than Gould had envisioned. I am one of them, studying evolutionary contingency at Michigan State University as part of a team working on the LTEE with E. coli, the longest-running and best-studied microbial experiment yet conducted.
Like human history, evolution plays out over time and involves a fundamental tension between chance and necessity.
Richard Lenski began the LTEE in 1988. For more than 66,500 generations, the project has followed the evolution of 12 initially identical populations of E. coli. These populations are grown in gently shaking flasks, incubated at body temperature, and filled with a broth that contains a small amount of glucose for food. Every day a researcher in the lab takes the flasks out of the incubator, transfers 1 percent of each population to fresh flasks of broth, and places them back in the incubator. The remaining 99 percent go into the refrigerator as a backup for two days before they are destroyed. Living samples of each population are frozen every 75 days, or about every 500 generations. Thus, ancestral and evolved clones can be revived for direct comparison and studied with other organisms at different stages of the experiment.
The experiment is highly simplified: The environment never changes. There is neither immigration nor emigration. Mutation is the sole source of the new variation that is grist for the mill of natural selection, to be retained if beneficial or purged if detrimental. No matter how beneficial it is, a mutation may also be lost at random because of the chance fluctuations of genetic drift—for instance, if its carrier were not in the lucky 1 percent of the population that is transferred to a new flask. This simplicity makes the experiment powerful, because mutation, natural selection, and genetic drift are the core processes of evolution that operate across all life.
The LTEE has essentially been a matter of rerunning evolution 12 times simultaneously. This “parallel replay” design allows the LTEE to examine how repeatable evolution is under identical conditions. But if the populations started the same, and conditions have been stable, how could they evolve differently?
The main reason is that mutations are spontaneous, random occurrences. Different mutations have arisen at different times and in different orders across the populations, giving each a unique mutational history. These differences can have consequences, because mutations can interact with one another, a phenomenon called epistasis. Each mutation can potentially change the effects and even the possibility of later mutations. Some mutations also change the ecology of the population, altering the conditions under which later evolution takes place. Mutations therefore open or close avenues of later evolution, much as events shape the flow of human history. Roosevelt survived Zangara’s attempt on his life, and that circumstance affected and caused later events. In exactly the same way, later evolution occurs in the context created by the evolution that has already happened.
When the LTEE began, it was known that the chance inherent to evolution’s core processes would likely cause the populations to have different evolutionary histories. The central question was whether or not those differences would yield meaningfully distinct outcomes—whether the populations would pursue parallel paths or tread divergent trails.
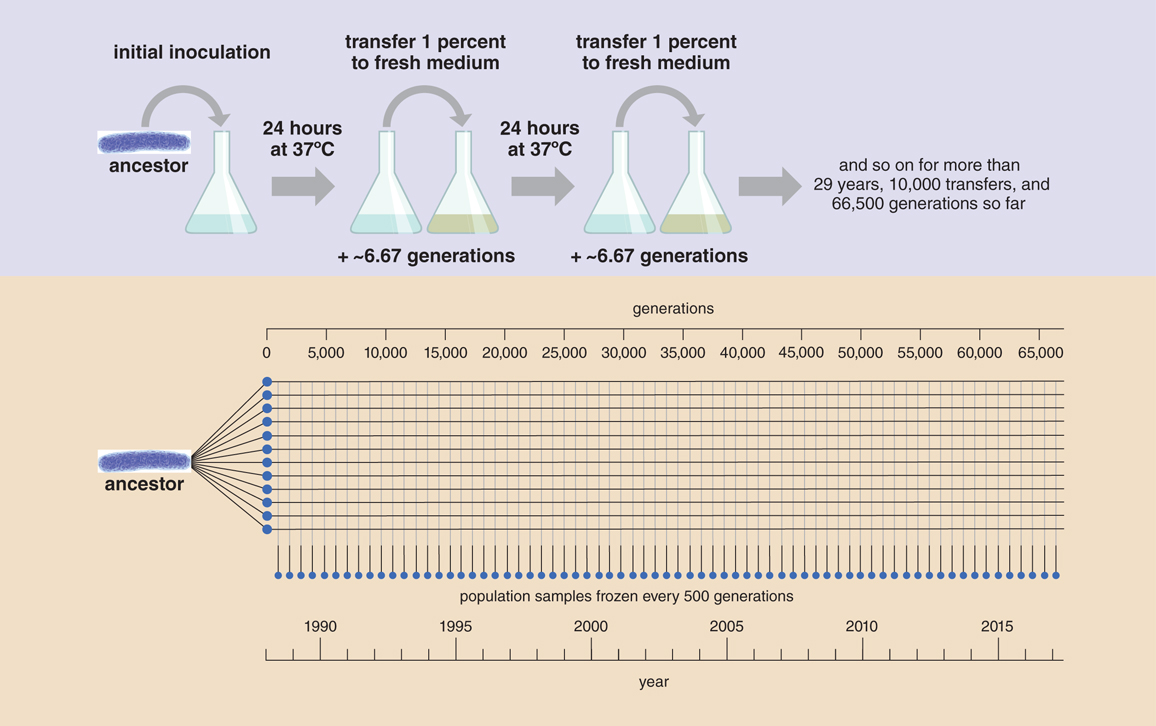
Illustration by Tom Dunne.
The LTEE populations have evolved in parallel in many ways. All have become much fitter in their environment through a common pattern of rapid early improvement that then slowed in pace but never stopped. By 60,000 generations, the populations had reached remarkably similar, but not identical, fitness levels. There have been other parallels, too: Adaptive mutations accumulated in a number of the same genes across the populations; all now are made up of larger cells that grow faster on glucose; and many have lost the ability to grow on certain substances they no longer encounter.
The populations have also diverged in a number of ways. Each population, of course, has its own distinct collection of mutations. Half of the populations have evolved defects in DNA repair, causing them to collect mutations much faster than the others. More impressively, a number of the populations have evolved simple ecosystems in which two or more lineages of cells coexist—a first step toward speciation. Most of these ecologies were discovered only in the past year using new technologies for sequencing the DNA of entire populations. We haven’t had a chance to investigate them, so we don’t know whether they are fundamentally distinct or essentially the same. These evolved differences generally have been subtle and require careful study to identify.
At a glance the populations all look pretty much alike. Some might argue that Conway Morris was right that contingency would only cause marginal differences between evolutionary replays. One population, however, bucked the trend and ostentatiously struck out on a different path. This population is incontrovertibly different from the rest, even to the naked eye. Its story has given the most valuable insights into contingency’s role in evolution to come out of the LTEE so far.
On a cold January day in 2003, more than 33,000 generations into the LTEE, transfer duty fell to Tim Cooper, a postdoc in the lab. As he checked the previous day’s flasks, 11 populations looked normal, like flasks of water with a drop or two of milk mixed in, only their slight cloudiness indicating the millions of resident bacteria. But one population, called Ara-3, was much cloudier, containing more cells than any other. A trait had evolved in the Ara-3 population that allowed the bacteria to use a new food source called citrate.
Citrate, the acid that gives orange juice its tang, has been in the LTEE from the beginning. It is a component of the population’s broth, not as food, but to help the bacteria acquire the iron that they need to thrive. Many bacteria can grow on citrate as a food source. E. coli can, too, but only when no oxygen is present. Unlike some of its close relatives such as Salmonella, however, E. coli cannot grow on citrate under the oxygen-rich conditions of the LTEE. Indeed, this “Cit–“ trait is part of what defines E. coli as a species. E. coli can’t produce a specialized protein called a transporter to bring citrate into the cell when oxygen is around. In essence, E. coli lacks a mouth for citrate under LTEE conditions. The LTEE bacteria were literally swimming in food they could not eat, missing the lemony dessert to their sweet meal of glucose. Lenski foresaw the possibility that the populations might one day evolve an aerobic, citrate-using, or “Cit+,” trait. But as the years rolled by, that trait seemed less likely to arise—until that extra cloudy flask came about, which turned out to be full of Cit+ E. coli cells.
The next year, in March 2004, I joined the lab as a graduate student and began to figure out why Cit+ took so long to evolve, and why it did so only once. Lenski and I thought there were two possible explanations: One was that the Ara-3 population had just gotten lucky. After all, Cit+ mutants of E. coli do happen, although they are vanishingly rare. (Only one was reported in the 20th century, and newer work has shown that obtaining the trait’s evolution usually requires long periods of intense selection utterly unlike what E. coli normally encounters in either the lab or nature.)
The other possibility had to do with contingency. What if Cit+ required multiple mutations to evolve? It would have been astronomically unlikely for all the mutations to occur at once, so they would have had to accumulate one by one over time. Ordinarily, natural selection can assemble mutations quickly if they build on each other by, for instance, improving a beneficial trait. But natural selection has no forethought and is blind to all but the fitness of an existing mutation. What if one or more were neutral? What if one or more were beneficial, but only under certain circumstances? If the Cit+ trait manifested only after all the needed mutations were present, then natural selection would not have been able to directly facilitate their accumulation, because the big reward came only at the end. Their assembly would have to be a chance outcome of evolutionary history and would likely take a long time. Under this hypothesis, Cit+ evolved in Ara-3 because one or more of the necessary mutations had accumulated during its unique history and “potentiated” Cit+ evolution by making the needed combination of mutations more likely.
If evolution is highly contingent, then it is inherently unrepeatable.
I tested these hypotheses exactly as Gould suggested: by replaying the tape of Ara-3’s evolution. To understand what I did, imagine in The Man in the High Castle that a time-traveling historian took her time machine back to different points in history to see whether the Allied victory in World War II depended on Roosevelt surviving the 1933 assassination attempt. If she were correct, then she would expect to see the Allies win far more often when she watched from points after Roosevelt survived. Although we obviously can’t conduct such an experiment with human history, we can do so in a laboratory evolution experiment. Indeed, what I did was similar, only without a time machine or insanely high stakes. I isolated dozens of frozen Cit– clones from different points in time, ranging from the original ancestor to right before Cooper’s discovery. I refounded the population over and over again with these clones, and reran evolution, looking for the pattern of reevolution of Cit+. If Cit+ had been historically contingent, then it would be more likely to reevolve after the “potentiating” mutations were in place. After all, if an outcome depends on a given event, such as a collection of mutations, then the outcome becomes more likely after that event.
These experiments kept me occupied for the better part of three years, during which I tested more than 40 trillion cells. I saw Cit+ reevolve 17 times, each time in replays founded with clones from 20,000 generations or later. Moreover, I demonstrated that later-generation clones have rates of mutation to Cit+ that are much higher than that of the ancestor. These results were perfectly in line with the contingency hypothesis and suggested that some mutation or mutations that had occurred by 20,000 generations had increased the potential for Cit+ to evolve. They showed that Ara-3 was not simply lucky enough to hit a jackpot any of its siblings could have gotten. Instead, its unique evolutionary history had mattered, because it allowed Ara-3 to eventually find the royal flush of mutations that produced Cit+ by making that hand more likely to occur.
I had shown that Cit+ evolution was historically contingent, but that left the questions of just what the underlying history was, what mutations constituted it, and how they contributed to the outcome. We knew that the history involved three phases: first, the accumulation of the “potentiating” mutations that increase the potential to evolve Cit+ in the LTEE; second, the occurrence of a final “actualizing” mutation that produced the switch from Cit– to Cit+; and third, the accumulation of mutations that “refined” the Cit+ trait. To figure out which mutations contributed to each phase, we had to go into the historical record of frozen samples, isolate clones from different time points, sequence them, and identify interesting mutations. In many ways, it was similar to the way a historian scours the historical record for evidence with which to identify events that might have led to an outcome of interest. Unlike the historian, when we found a mutation we thought might have been involved in Cit+ evolution, we were able to use genetic engineering techniques to directly test its effects. My colleagues and I have used this approach to figure out the history of Cit+ evolution in great detail. There is still more to plumb, but the following, much of which comes from the innovative work of a team led by Erik Quandt, who is now a postdoc at the University of Texas at Austin, is substantially complete.
Early in the history of Ara-3, a clone evolved that was able to compete better for scarce glucose by eating faster, but at the price of sloppiness. As it ate, it leaked a metabolic by-product into the broth called acetate, which is the acid found in vinegar. (Think of a messy toddler spilling half of his dinner on the floor.) After running out of the glucose, the cells would then turn to eating the acetate. The acetate presented an opportunity, however, and just before 20,000 generations, a mutant evolved that would switch to eating the acetate early. (Think of a younger toddler who leaves her plate early to eat her brother’s dropped food.) This mutant founded a new lineage of cells in the population, which was now made up of two coexisting cell types: sloppy glucose eaters and glucose-acetate users. A few thousand generations later, another mutation occurred in one of the glucose-acetate users. This mutation altered the tricarboxylic acid (TCA) cycle, the metabolic pathway by which cells process acetate, resulting in a mutant that was more specialized for growing on acetate. (Now a baby crawls over and eats the dropped food.) This mutant founded a third lineage in the population, which coexisted with the other two. Later, a third mutation occurred in an acetate-specialist that tweaked the TCA cycle again and made it even better at using the acetate. (The family dog realizes there is food on the floor and is even better at snatching it up.)
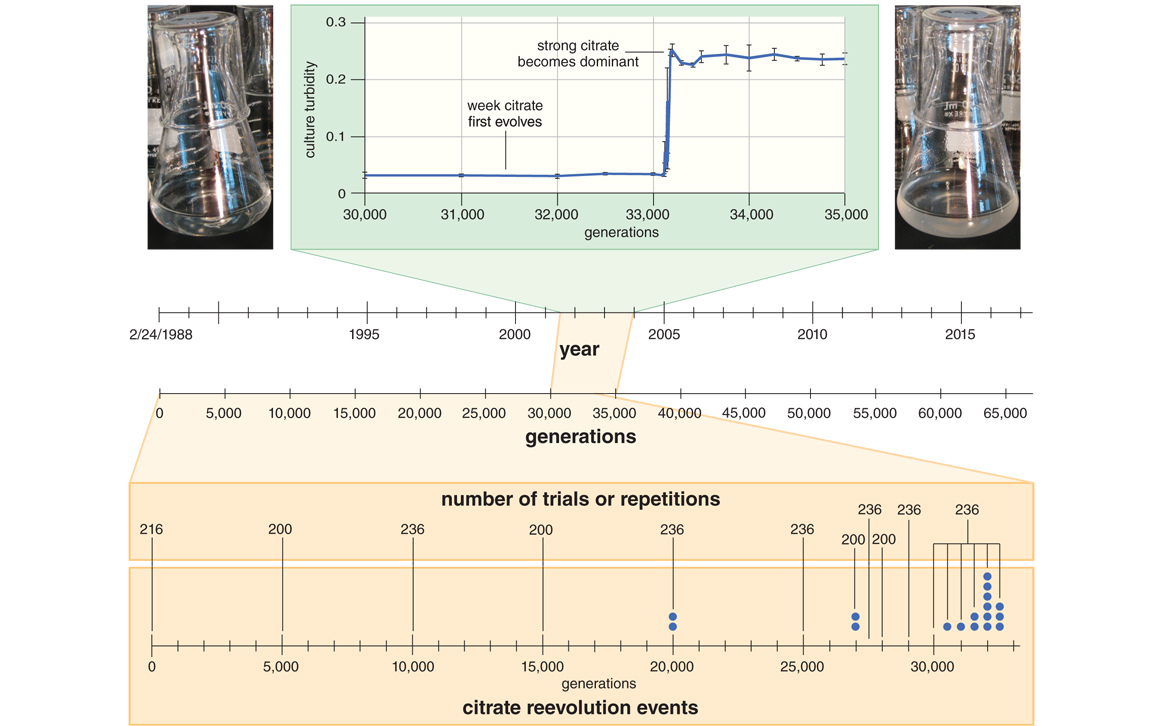
Illustration by Tom Dunne.
What does this have to do with citrate? As it happens, citrate is also metabolized via the TCA cycle, so the history of adaptation to growth on acetate coincidentally evolved cells that were ready to grow on citrate even before they could do so. The stage was set for an extraordinarily rare mutation that occurred in an acetate super-specialist cell at around 31,000 generations. This mutation was a duplication that placed two identical stretches of DNA side by side. In that duplicated DNA was a gene called citT, which encoded a protein that could transport citrate into the cell. It was part of a genetic instruction that told the cell, “Make citrate transporters only when there’s no oxygen around.” The duplication shuffled the preexisting genetic elements to make a new instruction that effectively read, “Make citrate transporters when oxygen is around.” This new instruction caused the switch from Cit– to weakly Cit+. Though weak, once the Cit+ trait existed, any new mutation that improved it was beneficial. The evolution of a type of E. coli that was good at growing aerobically on citrate was no longer historically contingent. Natural selection deterministically assembled mutations that refined the trait until Cit+ clones evolved that were strong enough to take over the population and cause the increased cloudiness that clued in everyone to the ongoing saga.
Was the emergence of an acetate ecology what led Ara-3 to take such a different evolutionary path? Not quite. Quandt found that some of the other populations had one or both of the first two sorts of acetate-adaptive mutations. It was the third mutation that set apart Ara-3. None of the other populations had it in addition to the other two mutations. (To continue the metaphor, none of the other populations seem to have gotten a dog.) Quandt showed that the third mutation had an important effect on the Cit+ trait. In early Cit+ clones, access to citrate was only slightly beneficial. But if the third acetate mutation were removed, the Cit+ trait would be harmful. Without that final tweak to the TCA cycle, access to the citrate throws a cell’s metabolism out of whack and makes it sick.
The other populations had taken different evolutionary paths that included acetate ecologies, but Ara-3 happened to take one that led it to develop a greater potential than the others to evolve Cit+. Due to this increased potential, evolving Cit+ in Ara-3 became more likely and repeatable after a certain point in time, as I showed in my replay experiments. The other populations’ histories did not include that point. We don’t know why Ara-3 took the path it did, and the others did not, but the difference mattered. Ara-3’s history leading to Cit+ was not bound to occur, but it did—and that, as Beatty has suggested, is the essence of evolutionary contingency.
The story of Cit+ shows how particular evolutionary histories that are not guaranteed to happen can coincidentally prepare the way for later innovations that probably wouldn’t evolve otherwise. Has contingency played this role in nature? Work done by Joseph Thornton and his colleagues at the University of Oregon and then the University of Chicago suggests that it very well might have.
Thornton’s team looked specifically at the evolution of a master regulator in bony vertebrates called the glucocorticoid receptor. It governs metabolism, stress and immune responses, and aspects of development. The key to the glucocorticoid receptor’s ability to play these roles is its specific sensitivity to the hormone cortisol. This evolutionary innovation sets apart the glucocorticoid receptor from its close relative, mineralocorticoid receptor, a regulator of salt concentration in the body that responds to cortisol and a few other hormones. Thornton and his crew aimed to figure out how the glucocorticoid receptor’s crucial specificity arose.
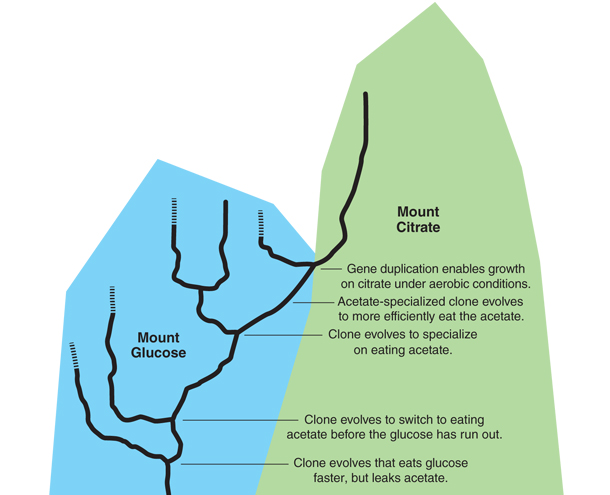
Illustration by Tom Dunne.
Led by Eric Ortlund of Emory University, Thornton’s team applied a technique called ancestral gene reconstruction to reconstruct the glucocorticoid receptor’s evolutionary history. They used computer programs to infer from the modern receptor and its relatives the sequences of three of its ancient ancestors. They then expressed these resurrected ancestral proteins in a modern organism and studied how their sequences, structures, and functions had changed during evolution. (They can’t know for certain that they are correct, of course; ancestral gene reconstruction studies are akin to how homicide investigators infer the details of a murder.) The earliest resurrected protein, which they called AncCR, from 450 million years ago, was ancestral to both the glucocorticoid and mineralocorticoid receptors and had the latter’s broad sensitivity. The next earliest protein, AncGR1, was resurrected from 440 million years ago and had similar sensitivity. The youngest, however, AncGR2, which was resurrected from 420 million years ago, had the glucocorticoid receptor’s trademark sensitivity to only cortisol. In other words, the glucocorticoid receptor’s ancestors evolved over the intervening 20 million years between AncGR1 and AncGR2 from hearing the “voices” of multiple hormones to hearing only that of cortisol.
Ortlund and his colleagues identified five mutations that had shifted the receptor’s sensitivity during that period, but there was a catch. When the researchers engineered a version of AncGR1 that had the five mutations, the result was a protein that was unresponsive to any hormone.
What was going on? The answer came when the team discovered that two other mutations that had occurred during this period were also important. The two mutations didn’t have any effect on hormone sensitivity at all by themselves. When they were added to the version of AncGR1 engineered to have the other five mutations, however, the result was a protein specifically sensitive to cortisol. Looking back to the transition from AncCR to AncGR1, the researchers found another mutation that likewise did not change sensitivity but was still critical to the later mutations’ effect. These three “permissive” mutations had allowed the five “sensitivity-shift” mutations to change the receptor’s specificity rather than lose its function. Similar to the “potentiating” mutations in the Cit+ story, the permissive mutations didn’t cause the innovative switch, but they increased the potential for it to occur.
Michael Harms, a postdoc in Thornton’s lab, later showed that the two subsequent permissive mutations were the only mutations that could have facilitated this pathway, given the receptor’s particular biophysical properties. We don’t know whether natural selection favored these mutations for some other reason or they were retained by chance, but we are certain that they were not favored for their effect on hormone sensitivity. With regard to the evolution of cortisol specificity, it was purely fortuitous that they stuck around for the millions of years needed for the five sensitivity-shifting mutations to accumulate. Had evolution taken another path, the glucocorticoid receptor would not have evolved because no potential for it to do so would have existed. After the permissive mutations evolved, however, the receptor’s evolution became more likely.
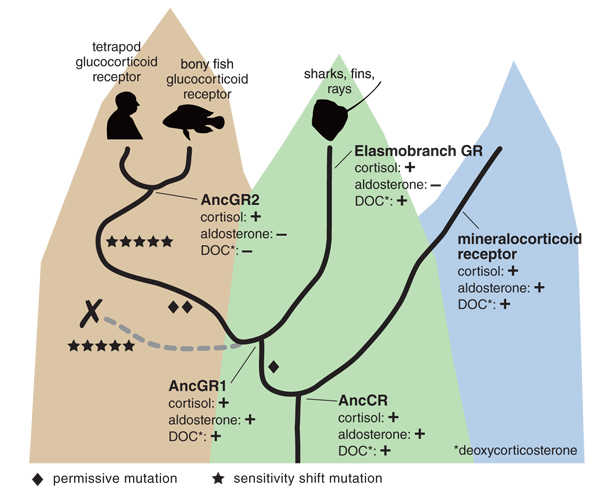
Illustration by Tom Dunne.
Thornton’s group showed that contingency in the evolution of a novel trait is not something restricted to the lab. Replay evolution again from 450 million years ago, and the glucocorticoid receptor might not evolve again. This prospect is significant because of the receptor’s many roles in bony-vertebrate metabolism and physiology. It is possible that creatures much like modern bony vertebrates would have evolved without the receptor, but the details of their physiology would have been different, with unknown ramifications. Perhaps more saliently, subtle changes in the developmental programs that the glucocorticoid receptor partly regulates have driven much of bony-vertebrate evolution. Strange as it may seem, the evolution of bony vertebrates, including the origin of humans, has been shaped by happenstance during the evolutionary changes in this single protein lineage. This realization is all the more staggering when one considers that this story is almost certainly not isolated. Future research will no doubt uncover other such stories.
Thornton’s group showed that contingency in the evolution of a novel trait is not something restricted to the lab. Replay evolution again from 450 million years ago, and the glucocorticoid receptor might not evolve again. This prospect is significant because of the receptor’s many roles in bony-vertebrate metabolism and physiology. It is possible that creatures much like modern bony vertebrates would have evolved without the receptor, but the details of their physiology would have been different, with unknown ramifications. Perhaps more saliently, subtle changes in the developmental programs that the glucocorticoid receptor partly regulates have driven much of bony-vertebrate evolution. Strange as it may seem, the evolution of bony vertebrates, including the origin of humans, has been shaped by happenstance during the evolutionary changes in this single protein lineage. This realization is all the more staggering when one considers that this story is almost certainly not isolated. Future research will no doubt uncover other such stories.
The study of historical contingency’s role in evolution is just beginning. Nevertheless, studies thus far have shed enough light to reveal some details of the landscape of evolutionary contingency. Indeed, those studies impart significant lessons that will inform future work and that I think point to a key role for evolutionary potential in determining the scope and effect of the contingency intrinsic to the evolutionary process.
One lesson is that Gould’s idea that evolution is completely unrepeatable is incorrect. Across the Greater Antilles, anole species with similar trait modifications that match comparable habitats evolved in a predictable fashion. Other biologists have identified similar instances of repeated ecomorph evolution among Hawaiian spiders, African cichlids, and Canadian sticklebacks. The anoles’ story is clearly not an oxymoronic one-off of repetition. Natural selection drives similar outcomes under similar conditions, showing that evolution can and does repeat itself.
The LTEE’s findings, on the other hand, show how such repeatability is not all pervasive. The 12 initially identical populations have evolved in strikingly parallel ways over the course of the experiment. But this convergence has not been unalloyed, because the populations have diverged as well. They display unique genetic, physiological, and ecological quirks beneath their similarities that show that each has happened onto a unique evolutionary path due to chance differences in their histories. One of the LTEE’s takeaways is that broad, general features such as evolutionary direction may be repeatable, but the details beneath them can vary due to historical happenstances. This lesson serves as a warning that convergence should not be overinterpreted because it can hide significant differences.
For example, my work on the Cit+ trait’s evolution demonstrates another lesson: Those contingent details can be quite important. The glucocorticoid-receptor evolution research shows that the importance of a particular history is not restricted to the lab, but also has mattered in the natural world. The scale of the glucocorticoid receptor’s history is breathtakingly grand, altering the trajectory of the lineages of humans and many other vertebrates. It is unclear how common this story is in protein evolution. Few proteins have had their molecular evolution worked out in similar detail so far. I suspect the glucocorticoid receptor’s story of functional shifts contingent upon historical happenstance is not uncommon.
The tales of both the Cit+ trait and the glucocorticoid receptor teach the lesson that history can matter in evolution. They also show how it matters by illuminating a key insight so glaringly obvious that we tend to forget it and its consequences: Evolution generally does not work from a blank slate, but from what exists. The deterministic part of evolution, natural selection, works by sorting through variation that arises. But that variation comes about through the mutation of an organism’s genotype, which is the product of its evolutionary history. In other words, history determines the “what” from which variation derives, and in turn determines an evolving lineage’s potential for future evolution, or “evolvability.”
Natural selection does, of course, play an important part in this history, but it is not an all-pervading one. After all, natural selection will favor a mutation if it provides an immediate benefit, but generally not if it simply increases the potential for beneficial variants to evolve down the road. Natural selection is not a chess player that can see several moves ahead. The epitome of opportunism, it sees only what is helpful or hurtful in the present moment. As a consequence, natural selection can speedily and deterministically assemble collections of immediately beneficial mutations, but how those collections affect later evolutionary potential will be largely coincidental. Hence, where evolution can go next is a contingent by-product of the details of this process and how it changes evolutionary potential by making unrealized traits or outcomes more or less likely.
Evolutionary potential holds promise as a means of approaching questions of evolutionary contingency and repeatability. For instance, evolutionary potential helps to explain how evolution can repeat itself while still being contingent. Evolutionary repetition will be more likely when prior history has either increased the potential for evolution to follow the adaptive path or paths leading to a given outcome or else has reduced the potential to go down other paths with different outcomes.
In the case of the anoles, for instance, the ancestral lizards that colonized the Caribbean had a prior evolutionary history that left them with physical traits that natural selection could easily tweak to repeatedly produce the same diversity of ecomorphs on each of the islands. Adaptation is more likely to recur if it requires only quantitative changes in existing traits—such as toe pad and body size, or leg and tail length—and the anoles had the right existing traits. The evolutionary potential for similar ecomorphs to arise, and hence for repetition and convergence, would no doubt have been less likely and common had the evolution of qualitatively new traits or body parts been necessary.
The anoles thus seem to have come onto the scene well-equipped to radiate, and their repeated radiations may have been virtually guaranteed. Other instances of repeated adaptive radiations may have involved ancestral organisms with similarly high intrinsic radiative potentials. If this idea is correct, repeated radiations could well be contingent events in that they depend on the existence of organisms with histories that have made them ready to radiate. Similar framing could also be applied to other occurrences of parallel evolution as well as to cases of convergent evolution. Indeed, this framing makes clear that evolutionary contingency does not mean that repetition never happens, although it may help explain why and under what conditions it does or does not happen.
Many aspects of the contingency intrinsic to the evolutionary process still need to be elucidated, including the conditions under which it may have the most or least significant effects and the levels at which it is important. (The same is, of course, true of the effects of contingency entering into the evolutionary process from outside, such as asteroid impacts and sudden climatological changes, which I did not examine.) If I am correct about evolutionary potential’s importance for understanding how history affects evolution, however, then developing a better grasp of evolutionary potential would be a good place to focus next. We still do not fully understand how to assess the evolutionary potential of an organism, much less how it might change and what constraints there might be on its manifestation. Evolvability is a complex phenomenon that involves genetic architecture, the process of mutation itself, how phenotype maps onto genotype, and how the range of accessible variation changes as a genotype evolves.
A better comprehension of evolutionary potential and its role in contingency will require a multidisciplinary effort involving not just evolutionary biologists, but also molecular biologists, geneticists, biophysicists, and systems biologists. Moreover, philosophers of science will be critical to this effort, because they can help scientists sharpen, parse, and explicate the concepts under examination. They would also be able to help formulate better hypotheses and design better, more incisive experiments that get at the core of these phenomena and how they interact.
In the end, we don’t yet know whether we’d get life remotely similar to what we have today if we reran evolution from primordial beginnings. Nor can we really predict what life on other planets might be like in any detail. Studies so far, however, strongly suggest that understanding evolutionary potential and how chance differences affect it would be a small but significant step toward addressing these questions. Identifying the conditions under which contingency becomes important to the evolutionary process will provide clues about how different outcomes might have been or might be elsewhere. Perhaps one day we will be able to know whether the living world was meant to be or if it was just one of many that could have been.
Click "American Scientist" to access home page
American Scientist Comments and Discussion
To discuss our articles or comment on them, please share them and tag American Scientist on social media platforms. Here are links to our profiles on Twitter, Facebook, and LinkedIn.
If we re-share your post, we will moderate comments/discussion following our comments policy.