Real-Deal Connectivity
By Catherine Clabby
New levels of detail in neural networks are being revealed
New levels of detail in neural networks are being revealed

DOI: 10.1511/2010.82.68
Until recently, images of neural circuits in mammals were based partly on statistical inference that was achieved by sampling some neurons and axons in one circuit. No more. Last winter, Ju Lu, a Harvard University graduate student in Jeff W. Lichtman’s laboratory, published the first complete map of a neuromuscular circuit—a connectome—in a mouse. The full picture revealed unexpected variation in the wiring of a complex nervous system. Lichtman explained its importance to American Scientist associate editor Catherine Clabby.
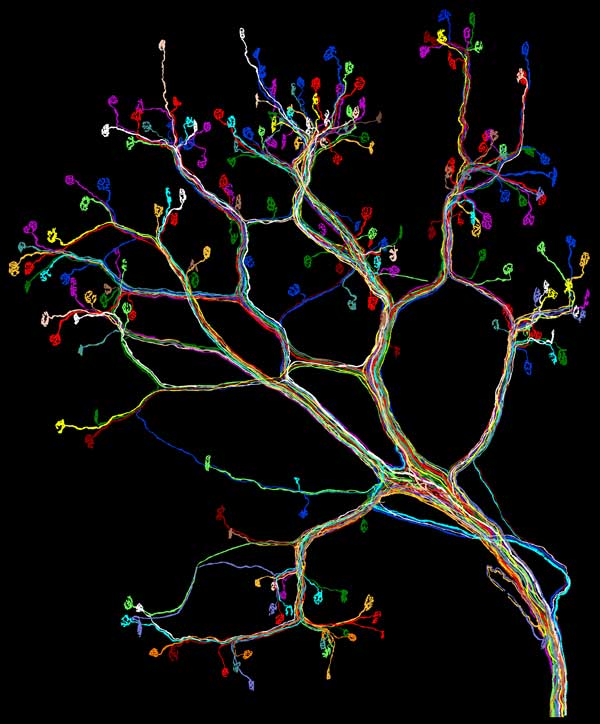
A.S. What prompted this project?
J.W.L. I’m a developmental neurobiologist. My research focuses on ways the nervous system goes from its developmental state to the fully functioning mature state. We think of the human brain as the most sophisticated and competent brain on the planet. We forget that at birth, remarkably, we have the most poorly functioning brain of any animal. A dragonfly with no training probably could chase a mosquito the first time it unfolds its wings. It takes much more time for humans to acquire and adjust to information about the world around us. For example, it takes a year to learn how to walk. Surprisingly, when you look at wiring diagrams of human and other mammalian brains in early postnatal development, their wiring is more complicated than it ends up in adulthood. Apparently, we trim away all but a small subset of neuronal interconnections initially established. Researchers generate these wiring diagrams, also known as connectomes, to understand what kind of rules shape the wiring, how it is that we encode our life experience into the wiring diagram of our brains.
A.S. When you saw them, what surprised you?
J.W.L. Nerves that move ears on either side of a mouse’s head are not exactly the same. That’s very different from the striking stereotypy that has been observed in animals such as insects. In invertebrates, genetic control over wiring ensures that each animal is optimally wired up in much the same pattern. But the mammalian nervous system seems a bit unfettered from genetic control. The guiding principle might be self-organization based on competitive interactions between nerve cells that begin by sharing the same targets but then vie for them. This group of nerve cells needs to come up with a solution to get a job done, that allows the function to occur (such as wiggling the ears) with no neuron taking on too much and no neuron doing not enough. We can see this process of self-organization unfold in muscles with time-lapse imaging. Initially several different nerves contact each muscle fiber. Due to competition, some branches are pruned while others persist. Self-organization may allow the nerve cells to create a workable solution that, while not necessarily optimized, is good enough. That gives rise to variation.
A.S. What technology makes this view possible?
J.W.L. We used genetically modified animals, altered to express fluorescent proteins in their spinal motor neurons. For this study we had to see every single nerve projecting to a muscle. In this case, every cell fluoresced yellow, but the wires intermingled into a tangled mass not unlike a bowl of yellow spaghetti. In order to trace out each one, we had to generate very good optical images. Then we had to very precisely trace each of the little wires. In this case we are talking about a trivially small number of motor neurons supplying this small muscle: just 15 axons reaching about 200 muscle fibers. But getting all the wires traced required tens of thousands of microscopy images and a data set that often approached 100 gigabytes for one muscle. The tracing was computer assisted but it was not fully automated. Ju Lu, who led this work, had to help the computer around the rough spots. It was slow. The first took six months to trace. Now things take a few weeks but it is still tedious work.
A.S. Where will this take your research?
J.W.L. I’d like to extend this kind of analysis to other parts of the nervous system during early development. We think doing this fluorescence imaging along with electron microscopy, which has better resolution, offers us a way of seeing every synapse in the brain of an animal of any age. When you start looking at younger animals, there are a lot more wires. They are thinner and packed closer together. Because we have developed means of using multiple colors to make “brainbow” brain images, we are hopeful that circuit tracing will get easier. For example, in developing muscles we can match the one purple axon entering a muscle with all the synapses in that same color without having to trace out every branch point. In young mice, we are beginning to see patterns in the ways nerves interact that we hadn’t seen before. These patterns show the power of tools that allow one to see things that were previously hidden. Many times we see things we didn’t expect—and that is good!
Click "American Scientist" to access home page
American Scientist Comments and Discussion
To discuss our articles or comment on them, please share them and tag American Scientist on social media platforms. Here are links to our profiles on Twitter, Facebook, and LinkedIn.
If we re-share your post, we will moderate comments/discussion following our comments policy.