Ultimate Resolution
By Catherine Clabby
Imaging specialists such as Richard Parton at the University of Oxford are breaking through the diffraction limit to get a better view of biology.
Imaging specialists such as Richard Parton at the University of Oxford are breaking through the diffraction limit to get a better view of biology.

DOI: 10.1511/2013.105.418
Until recently the resolution of optical microscopy was limited by something considered insurmountable: the diffraction limit. Light spreads out when passing through a small aperture, producing a blurry focal spot. That makes any details smaller than about 200 nanometers wide—including many of the fundamental structures of living cells—look fuzzy. But new strategies, hardware, and software have shattered that limit. A technology called structured illumination microscopy, or SIM, can reach an astonishing resolution of 100 nanometers in cells tagged with fluorescent dyes. SIM manipulates both the pattern of illumination and the timing of when the dye tags glow. American Scientist contributing editor Catherine Clabby spoke with Richard Parton at Micron Oxford about the groundbreaking images he and others have made with the Delta OMX Blaze SIM. The microscopes are improving the view in labs around the world.
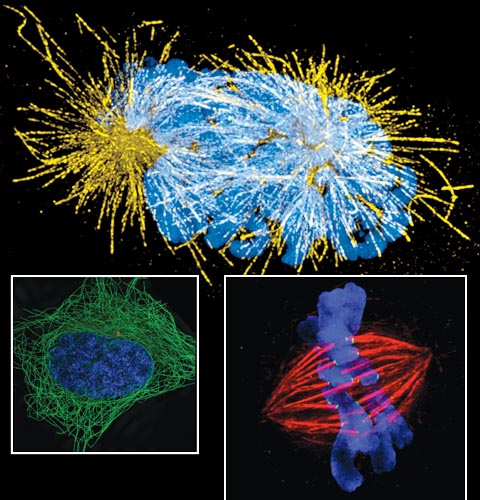
Images courtesy of Markus Posch,University of Dundee; Jane Stout, Indiana University; and Steffen Lawo, University of Toronto
What are people seeing with SIM technology that could not be seen before?
Researchers from diverse fields are seeing many things that are fundamental to normal cell activity. Scientists can better observe activity from the subtleties of DNA organization inside nuclei during genome replication to the gene expression in mammalian systems. This resolution makes it possible to capture how the organization of DNA in chromosomes relates to function during gene expression, DNA replication, and repair. A finer view of structures such as centrosomes during mitosis are now possible, for example. When they function properly, centrosomes promote chromosome stability. Understanding how all these functions work normally gives insight into what might occur when they malfunction and contribute to human disease.
What specifically are you studying in your laboratory using this technique?
While at Imperial College, Alice Brown and Dan Davis collaborated with our research group, run by cell biologist Ilan Davis. They probed the long-standing question of how natural killer cells delivered lytic granules to their targets. It was known that a dynamic actin mesh underlies the cell membrane but conventional resolution light microscopy was not able to resolve its organization. Using two-color, three-dimensional SIM allowed Brown to characterize the pore size of the actin mesh and the distribution of lytic granules. That revealed how these granules specifically associate with open regions of actin mesh in stimulated cells. This remodeling is likely important in other types of secretion as well.
At Micron Oxford, Tim Weil and I investigated the mechanisms of setting up the body plan of an organism during development—known as developmental patterning—in fruit fly eggs. It is understood that particular proteins need to be expressed at particular times and locations to define the anterior and posterior of an organism. It has also been shown that this localized protein expression, in many cases, is determined by the localization of messenger RNAs (mRNAs). We were able to determine the dynamics of mRNA transport particles in relation to a particular type of subcellular components known as processing bodies. With fast and live imaging supported by 3D SIM and electron microscopy, we saw the ways that important mRNAs with separate translational fates interact differentially with those structures. That could help explain how developmental patterning occurs.
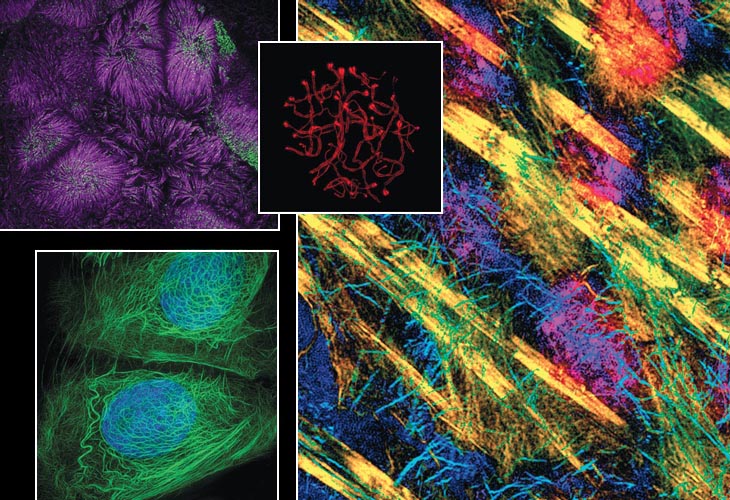
Image at top left courtesy of Matthew Tyska, Vanderbilt University; right, courtesy of Tim Weil, University of Oxford. Images at upper middle and bottom left courtesy of Graham Wright, Institute of Medical Biology.
What are your biggest challenges?
John Sedat of the University of California, San Francisco, and the other SIM inventors gave science a new means to explore a wide range of biological questions. As with all imaging applications, however, detector sensitivity, speed, and the brightness and photo stability of fluorescent labels limit what can be achieved. They determine the amount of light that is available to generate an image. Many labs around the world are striving to make improvements.
In collaboration with Christian Eggeling at the Weatherall Institute of Molecular Medicine at Oxford, our group is trying to take the resolution of live imaging down to 50 nanometers or better. We’d like to take advantage of the way some fluorescent probes switch between fluorescent and nonfluorescent states and combine this with structured illumination to effectively go far below the diffraction limit.
Where could this technology go next?
Together with the lab of Martin Booth at the University of Oxford, we are also investigating the use of what is called adaptive optics approaches by manipulating the light rays that make an image. We want to improve our ability to image rapidly, by developing a fast-focus method, and to correct for distortions that deteriorate image quality when we are imaging inside a cellular structure.
Another common limitation in this kind of imaging is getting the right dyes into living biological material. We’re teaming up with chemists and biochemists, including Mark Howarth at Oxford. He is working on more effective alternatives to conventional peptide tags or antibody labeling for endogenous proteins. The next challenge we see for the OMX platform is to push structured illumination toward the ultimate goal of unlimited resolution in living biological material.
Click "American Scientist" to access home page
American Scientist Comments and Discussion
To discuss our articles or comment on them, please share them and tag American Scientist on social media platforms. Here are links to our profiles on Twitter, Facebook, and LinkedIn.
If we re-share your post, we will moderate comments/discussion following our comments policy.