In Living Color
By Felice Frankel
A new technique for "labeling" neurons with brilliant hues reveals structures and connections in the brains of transgenic mice
A new technique for "labeling" neurons with brilliant hues reveals structures and connections in the brains of transgenic mice

DOI: 10.1511/2008.69.59
"Sightings" often profiles images that receive well-deserved public attention. In this issue, we sample a group of images recently featured on the cover of Nature and in newspapers, magazines and science Web sites around the world. These colorful images are wonderfully compelling and, more important, highly informational. Some clever thinking and fine biology has gone into developing a stunning new technique for "labeling" neurons to reveal structures and connections in the brain. We interviewed Jeff W. Lichtman (J. W. L.), professor of molecular and cellular biology at Harvard University, and Jean Livet, a postdoctoral fellow in his lab.
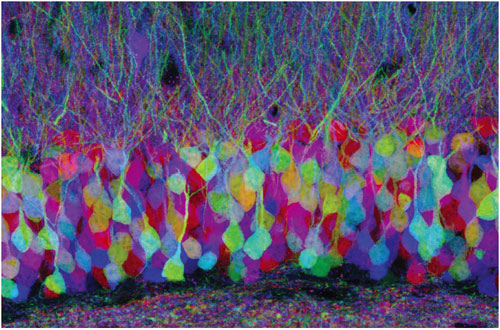
Jean Livet, Harvard University
F. F. Jeff, tell us how these colors were created.
J. W. L. The colors in these images come from jellyfish and coral genes that encode proteins that are fluorescent—proteins that glow particular colors (red, green and blue) when light of the appropriate wavelengths is shone on them. These genes obviously don't belong in mice. Biologists have used staining to see nerve cells and their connections for a long time—ever since 1887, when Santiago Ramón y Cajal applied Camillo Golgi's silver stain. But when all neurons carry the same stain, it's hard to distinguish them. Our previous experiments have shown that we can insert genes coding for different-colored fluorescent "stains" into transgenic mice in a permanent way.
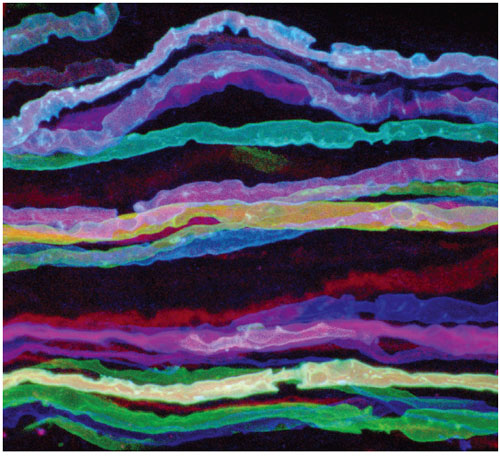
Jean Livet, Harvard University. Republished by permission from Livet et al., Nature 450:56–61
Josh Sanes and I previously collaborated to create mice that express pure red, pure yellow, or pure blue fluorescence in their neurons. But the only way we could get more than one color was to cross these mouse lines, and that did not cause much variability. In order to get more colors and thus better distinguish cells, we needed a strategy to randomize the amount of red, green and blue fluorescent protein in each nerve cell.
Jean Livet, a postdoctoral fellow in my laboratory, thought long and hard about this and came up with an ingenious idea: Create a random choice of expression using DNA recombination. He built a transgene bearing three differently colored fluorescent-protein genes, in which recombination would randomly switch on the expression of one of the fluorescent proteins. The trick is that if the nerve cell has multiple copies of this transgene, it will play this game of chance multiple times, ending up with a randomized mixture of red, green and blue. By analogy, one can think of a slot machine: When you pull the lever, you set off the recombination. If you end up with two lemons and a cherry, the color combination appears as light orange. This transgenic slot machine probably has closer to 10 copies of the transgene than three—giving a large number of possible combinations.
F. F. Jean, do you remember how you came up with the idea? What was the train of thought bringing you to this novel approach?
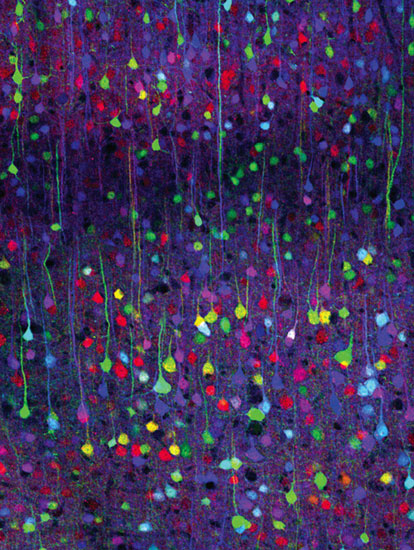
Tamily A. Weissman, Harvard University
J. L. After I joined the Lichtman lab in 2002, Jeff and his collaborator Josh Sanes and I did some brainstorming about how to solve problems in seeing nerve cells. Over several weeks, Jeff kept repeating this comment: "More colors would be helpful." I was dreaming of a mouse in which all the motor axons would be different colors, but that did not seem possible. In the Sanes lab, Guoping Feng and Mario Buffelli had created mice whose neurons glowed only when a sequence suppressing gene expression was removed by a DNA-recombination system called Cre/lox. The enzyme Cre (a recombinase) can switch gene expression on by removing or inverting sections of DNA flanked by a pair of sequences called lox sequences.
I thought about the possibility of trying a scheme that was already used in gene-targeting technology, where this recombination system can create two kinds of mutant strains from the same genetic material. In this scheme, more than two lox sequences are inserted within a single construct, giving Cre a "choice" of locations where it can act to catalyze recombination.
Jeff immediately pointed out that Cre would certainly recombine the two extreme sites of the lox array and excise the whole construct. How to stop this? It popped into my mind that incompatible lox variant sequences (which were increasingly in fashion for cloning at that time) could be used for that purpose.
I talked to my friend Hiroshi Nishimune, then a postdoc with Josh Sanes and now an assistant professor at the University of Kansas School of Medicine. Hiroshi confirmed that the idea could perhaps work. Jeff and Josh, coming out of Josh's office, joined our discussion, and the name "Brainbow" was proposed for our recombinatorial color scheme—I believe by Jeff. Once we had that name, we had to do it! We spent about three weeks scribbling DNA constructs, using little triangles for loxsites. Josh was instrumental in the key process of deciding the final form and details of the construct. During these brainstorming weeks I came up with a second idea, creating multiple recombination choices using DNA inversion. Then I spent about a year just making the constructs and testing them. It's ironic that my objective at the start of my postdoc was to do as little molecular biology as possible and concentrate on imaging!
Click "American Scientist" to access home page
American Scientist Comments and Discussion
To discuss our articles or comment on them, please share them and tag American Scientist on social media platforms. Here are links to our profiles on Twitter, Facebook, and LinkedIn.
If we re-share your post, we will moderate comments/discussion following our comments policy.