Brain Surgery Without a Knife
By Joseph Wilson
Focused ultrasound shows promise as a treatment for neurological conditions such as Alzheimer’s disease and addiction.
Focused ultrasound shows promise as a treatment for neurological conditions such as Alzheimer’s disease and addiction.

DOI: 10.1511/2025.113.1.4
There are few words that a patient wants to hear less than “brain surgery,” but for a wide range of neurological diseases, surgery remains the only treatment option. But what about surgery without a knife?
A therapeutic technology called focused ultrasound (FUS) shows promise as a nonsurgical alternative to brain surgery for some ailments. In recent years, neurosurgeons have been using FUS to treat essential tremor disorder and Parkinson’s disease. It is also being investigated as a way to treat a diverse range of neurological conditions including Alzheimer’s disease, drug addiction, and even brain tumors, all without invasive surgery.
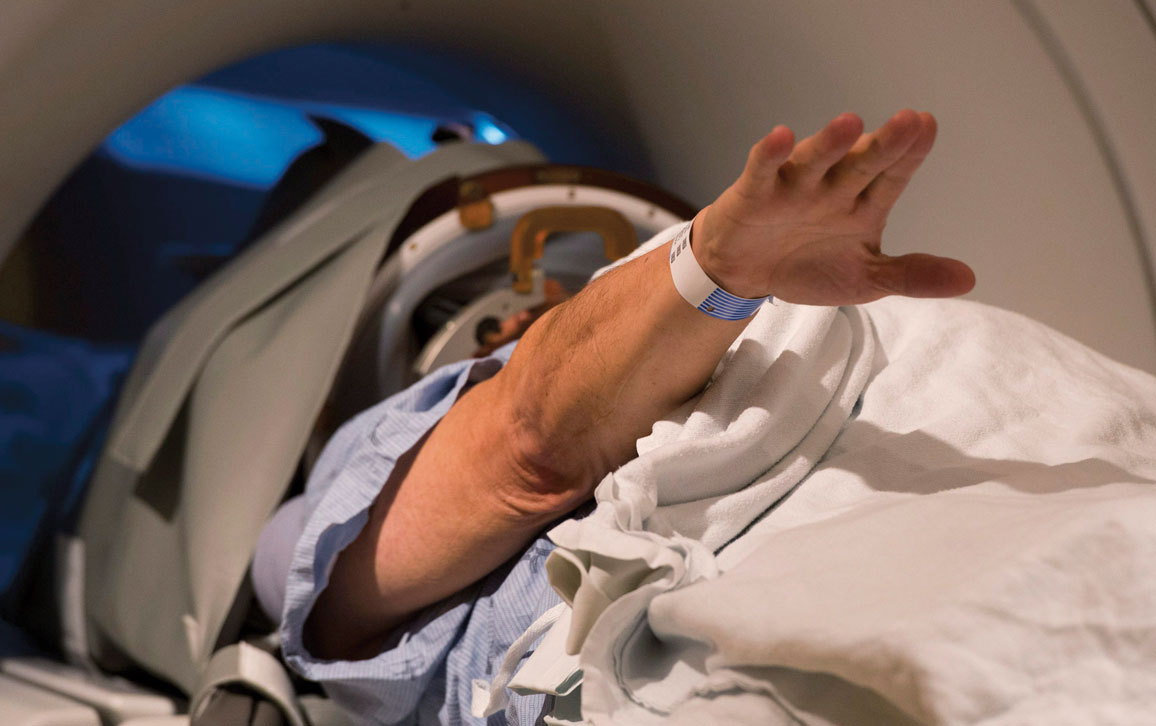
AP Photo/The Canadian Press, Frank Gunn
Patients who opt for FUS undergo magnetic resonance imaging (MRI) while wearing a special helmet that is attached to 1,024 ultrasound probes. “The sound waves are tuned so that they converge at precise locations in the brain,” says Ali Rezai, the director of the Rockefeller Neuroscience Institute at West Virginia University. Where the waves converge, sonic energy is converted into heat that kills cells in a very specific location, as little as a millimeter wide. When treating patients with essential tremor or Parkinson’s disease, “we can modulate the dosage or increase the energy to create a small thermal lesion to stop tremors,” Rezai says.
For some patients, FUS can provide an incision-free alternative to deep brain stimulation (DBS), a common surgical procedure used to treat neurological disorders such as Parkinson’s disease and essential tremor, as well as epilepsy and obsessive-compulsive disorder. DBS patients have electrodes surgically implanted directly into their brains. The electrodes, which are controlled by a pacemaker-like device implanted near the patient’s collar bone, stimulate brain cells through a form of neuromodulation. The treatment works well, but it involves invasive surgery. “FUS allows patients to have a choice,” says Vibhor Krishna, a neurosurgeon at the University of North Carolina at Chapel Hill. “There are patients who would choose DBS, and then there are patients who would never choose DBS. Today, we have an option for those patients.”
Researchers are still trying to understand the neurological mechanisms that make treatments such as FUS and DBS effective; nonetheless, the results of these procedures can be astounding. Rezai shows me a video of one of his patients with essential tremor before an FUS procedure. “You can see that he has severe shaking of his arm and legs, and he has severe difficulty doing basic activities: writing, eating, drinking, brushing his teeth,” Rezai says. “He gets the ultrasound with increased energy to the part of the brain causing the tremor, and we shut it down.” In a postprocedure video, the tremors have vanished. The entire process takes only a couple of hours, and, according to Rezai, for many of the patients the tremors never return.
FUS could also potentially be used to deliver medications to the brain with greater efficacy. Medicating the brain has been a challenge for neuroscientists because of the blood–brain barrier, which separates neural blood vessels from surrounding brain tissue. Large, harmful molecules are stopped from passing from the blood to the brain, but therapeutic drugs are also swept up in the dragnet. For example, Alzheimer’s disease medications such as aducanumab and lecanemab work by dissolving brain plaques that interfere with communication pathways. However, the blood–brain barrier limits these drugs from reaching their targets.
“A basic 20-minute neuromodulation treatment can result in sustained reduction in cravings and drug use more than three months after the procedure.”
In January 2024, Rezai and his team published a paper in the New England Journal of Medicine that showed how FUS could be used to allow aducanumab and other medications to pass through the blood–brain barrier unencumbered. FUS agitates microbubbles that are injected intravenously. These bubbles expand and create gaps in the blood–brain barrier through which the drugs can pass. The team’s study of administering aducanumab with FUS showed a 50-percent reduction in brain plaque coverage in the treated areas. “The temporary opening of the blood–brain barrier by focused ultrasound allows more of the antibody to get into the brain,” Rezai explains.
Rezai is also looking to FUS therapies for treating different types of addiction through neuromodulation. Low levels of ultrasound energy are used to stimulate brain cells, but. unlike DBS, there is no need for an electrode implant. Preliminary results were published in the journal Frontiers in Psychiatry in 2023 as part of an ongoing study with the U.S. National Institutes of Health and the National Institute on Drug Abuse.
Rezai’s hope is that FUS treatment “can reset parts of the brain that are focused on addiction or mental health problems without creating a lesion. Drugs, alcohol, gambling—it doesn’t matter. It’s the same part of the brain that is electrically supersensitive. We deliver ultrasound waves to calm the hypersensitivity of that part of the brain, calming the neurons.” Although Rezai’s team has published results from only four patients, “what we’ve seen is a basic 20-minute neuromodulation treatment can result in sustained reduction in cravings and drug use more than three months after the procedure,” he says.
Substance use disorder and other types of addiction are more complex biopsychosocial phenomena than are the tremors brought on by Parkinson’s disease. Rezai is emphatic that for people suffering from addictions and other mental health issues, technologies such as FUS will need to be accompanied by therapy and broader social supports.
“It’s the only way,” he says. “[FUS] is an adjunctive approach that empowers the therapist to do talk therapy and prescribe medications because the brain has been reset. The patients are more engaged in their treatment plan and more open to the therapist making an impact.”
Neurosurgeons are continuing to investigate other potential uses of FUS. Krishna is currently involved in phase I clinical trials for the use of FUS for treatment-resistant epilepsy. He tells me about another trial at Sunnybrook Hospital in Toronto that used FUS on patients with brain tumors. “You open the blood–brain barrier and pair that with chemotherapy; it allows you to then treat it in a targeted fashion,” Krishna says.
The opening of the blood–brain barrier also allows for a procedure called a liquid biopsy. “We could biopsy any material that leaks out of the tumors into the patient’s blood,” Krishna says. “We could diagnose tumors, but then do surveillance of the tumors to see how they respond to treatment.” Rezai is pursuing a similar line of inquiry in neurodegenerative conditions. “You could sample antigens or molecules coming from the brain into the blood and get a blood test to phenotype or tissue-type a tumor without doing a surgical biopsy,” he says.
This kind of procedure would certainly be welcome news to those facing the prospect of brain surgery. “It’s surgery on the inside even though there’s no incision on the outside,” Krishna says.
Click "American Scientist" to access home page
American Scientist Comments and Discussion
To discuss our articles or comment on them, please share them and tag American Scientist on social media platforms. Here are links to our profiles on Twitter, Facebook, and LinkedIn.
If we re-share your post, we will moderate comments/discussion following our comments policy.