
This Article From Issue
March-April 2017
Volume 105, Number 2
Page 75
In this roundup, digital features editor Katie L. Burke summarizes notable recent developments in scientific research, selected from reports compiled in the free electronic newsletter Sigma Xi SmartBrief. Online: https://www.smartbrief.com/sigmaxi/index.jsp
Moon Is Older Than Expected
Lunar rocks and soil collected in 1971 during the Apollo 14 mission have been analyzed to show that the Moon is older than expected, 4.51 billion years old. Earlier estimates had postulated that the Moon formed within 100 million to 200 million years after the Solar System formed, but the study put it at about 60 million years after that event. Tiny bits of the mineral zircon were extracted from the lunar material and their age was estimated using uranium-lead radioisotope dating. This result indicates that the Moon-forming impact occurred much sooner after Earth’s formation, and also that Earth could have cooled down enough to allow life earlier than previously thought as well.

NASA
Barboni, M., et al. Early formation of the moon 4.51 billion years ago. Science Advances doi:10.1126/sciadv.1602365 (January 11)
Self-Assembling Nanowires
A team led by Nicholas Melosh of Stanford University has created the tiniest wires ever made: conductive cables just three atoms wide composed of interlocking cages of carbon and hydrogen known as diamondoids. The researchers’ technique could be used to build nanowires for many applications, such as fabrics that act as generators, optical devices, and materials that conduct electricity without any loss. The nanowires are novel in that they self-assemble with a solid, crystalline core capable of conducting electricity. This semiconducting core, made with a combination of copper and sulfur, is surrounded by the diamondoids, which form an insulating shell. The group has already made these one-dimensional nanowires with cores of cadmium, zinc, iron, and silver, each of which can work with different materials and can have various applications.
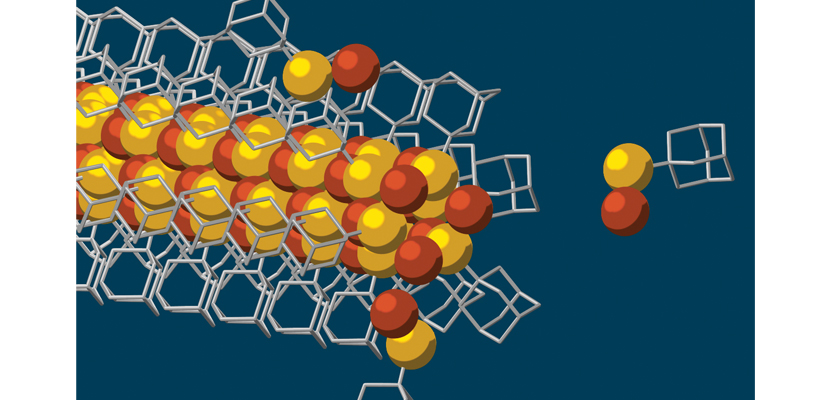
SLAC National Accelerator Laboratory
Yan, H., et al. Hybrid metal-organic chalcogenide nanowires with electrically conductive inorganic core through diamondoid-directed assembly. Nature Materials DOI: 10.1038/nmat4823 (December 26)
Cells with Semisynthetic DNA
Bacteria with a genetic code of six nucleotides, the building blocks of DNA, were engineered successfully. DNA in nature is made up of four nucleotides. Previous research had shown that synthetic nucleotides designated as X and Y could be incorporated into Escherichia coli bacteria, but that the engineered microbes grew slowly and expunged the foreign nucleotides after several generations. This new study showed that the engineered bacteria could retain the synthetic nucleotides over 60 divisions. The key to this progress was a change to a molecular transporter, which helps the bacteria import the unnatural nucleotides. Synthetic nucleotides such as these could allow more directed engineering of bacteria.
Zhang, Y., et al. A semisynthetic organism engineered for the stable expansion of the genetic alphabet. Proceedings of the National Academy of Sciences of the U.S.A. doi:10.1073/pnas.1616443114 (January 23)
Dinosaur Eggs Slow to Hatch

AMNH/M. Ellison
Many recent discoveries about dinosaurs have highlighted how many more traits they share with birds than we once thought, but a new study notes a trait that is decidedly reptilian: lengthy egg incubation. Developing dinosaur embryos incubated for 2.8 to 5.8 months, far longer than those of birds with eggs of similar size, which take 45 to 80 days. Fossilized dinosaur embryos are a rare find in paleontology, but the team used specimens from two species, Protoceratops andrewsi and Hypacrosaurus stebingeri. Counts of daily growth lines in the teeth of the fossilized dinosaur embryos suggested a development period closer to that of reptiles and about double what one would expect for a bird egg of similar size. These longer incubation times could have contributed to the dinosaurs’ vulnerability at the end of the Cretaceous.
Erickson, G. M., D. K. Zelenitsky, D. I. Kay, and M. A. Norell. Dinosaur incubation periods directly determined from growth-line counts in embryonic teeth show reptilian-grade development. Proceedings of the National Academy of Sciences of the U.S.A. 114:540–545 (January 17)
Prions Found in Bacteria
Once thought to occur only in eukaryotic cells, prions—infectious misfolded proteins responsible for illnesses such as “mad cow” disease—have now been observed in bacteria. A section of protein from the bacterium Clostridium botulinum acts like a prion when inserted into yeast or Escherichia coli bacteria. Researchers analyzed about 60,000 bacterial genomes to search for prion-forming proteins in yeast, homing in on a section that regulates the activity of many genes. The potentially prion-forming section of protein from C. botulinum was inserted into E. coli and yeast. In the E. coli, clumps of the protein aggregated, a characteristic of prions. In the yeast, the section of protein could replace the functions of another prion-forming protein. In this instance, many genes were active that were suppressed by the normal version of that protein. This finding suggests that prions could be adaptive in certain environments. Because prions are heritable, bacteria could pass along traits without a genetic mutation. Next, the researchers want to find out whether the section of protein they isolated acts like a prion in its original host—a task that may be challenging, because C. botulinum is difficult to work with in the lab. Studying prions in bacteria could also lead to important breakthroughs in understanding the behavior of prions in human diseases.
Yuan, A. H., and A. Hochschild. A bacterial global regulator forms a prion. Science 355:198–201 (January 13)

American Scientist Comments and Discussion
To discuss our articles or comment on them, please share them and tag American Scientist on social media platforms. Here are links to our profiles on Twitter, Facebook, and LinkedIn.
If we re-share your post, we will moderate comments/discussion following our comments policy.